[English] 日本語
 Yorodumi
Yorodumi- EMDB-30465: Cryo-EM structure of dengue virus serotype 2 in complex with the ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30465 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of dengue virus serotype 2 in complex with the scFv fragment of the broadly neutralizing antibody EDE1 C10 | ||||||||||||
 Map data Map data | Dengue 2 NGC With ScFv C10 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Dengue virus / antibody / broadly neutralizing / flavivirus / VIRUS | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / host cell mitochondrion / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / ribonucleoside triphosphate phosphatase activity / viral capsid / double-stranded RNA binding / channel activity / monoatomic ion transmembrane transport / clathrin-dependent endocytosis of virus by host cell ...symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / host cell mitochondrion / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / ribonucleoside triphosphate phosphatase activity / viral capsid / double-stranded RNA binding / channel activity / monoatomic ion transmembrane transport / clathrin-dependent endocytosis of virus by host cell / mRNA (nucleoside-2'-O-)-methyltransferase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / protein dimerization activity / host cell endoplasmic reticulum membrane / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / induction by virus of host autophagy / viral RNA genome replication / serine-type endopeptidase activity / RNA-dependent RNA polymerase activity / virus-mediated perturbation of host defense response / fusion of virus membrane with host endosome membrane / viral envelope / host cell nucleus / virion attachment to host cell / structural molecule activity / virion membrane / proteolysis / extracellular region / ATP binding / membrane / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Dengue virus 2 / Dengue virus 2 /  Homo sapiens (human) / Homo sapiens (human) /   Dengue virus Dengue virus | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||
 Authors Authors | Zhang X / Sharma A | ||||||||||||
| Funding support |  France, France,  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses. Authors: Arvind Sharma / Xiaokang Zhang / Wanwisa Dejnirattisai / Xinghong Dai / Danyang Gong / Wiyada Wongwiwat / Stéphane Duquerroy / Alexander Rouvinski / Marie-Christine Vaney / Pablo Guardado- ...Authors: Arvind Sharma / Xiaokang Zhang / Wanwisa Dejnirattisai / Xinghong Dai / Danyang Gong / Wiyada Wongwiwat / Stéphane Duquerroy / Alexander Rouvinski / Marie-Christine Vaney / Pablo Guardado-Calvo / Ahmed Haouz / Patrick England / Ren Sun / Z Hong Zhou / Juthathip Mongkolsapaya / Gavin R Screaton / Felix A Rey /      Abstract: The human monoclonal antibody C10 exhibits extraordinary cross-reactivity, potently neutralizing Zika virus (ZIKV) and the four serotypes of dengue virus (DENV1-DENV4). Here we describe a comparative ...The human monoclonal antibody C10 exhibits extraordinary cross-reactivity, potently neutralizing Zika virus (ZIKV) and the four serotypes of dengue virus (DENV1-DENV4). Here we describe a comparative structure-function analysis of C10 bound to the envelope (E) protein dimers of the five viruses it neutralizes. We demonstrate that the C10 Fab has high affinity for ZIKV and DENV1 but not for DENV2, DENV3, and DENV4. We further show that the C10 interaction with the latter viruses requires an E protein conformational landscape that limits binding to only one of the three independent epitopes per virion. This limited affinity is nevertheless counterbalanced by the particle's icosahedral organization, which allows two different dimers to be reached by both Fab arms of a C10 immunoglobulin. The epitopes' geometric distribution thus confers C10 its exceptional neutralization breadth. Our results highlight the importance not only of paratope/epitope complementarity but also the topological distribution for epitope-focused vaccine design. #1:  Journal: Nature / Year: 2016 Journal: Nature / Year: 2016Title: Structural basis of potent Zika-dengue virus antibody cross-neutralization. Authors: Barba-Spaeth G / Dejnirattisai W / Rouvinski A / Vaney MC / Medits I / Sharma A / Simon-Loriere E / Sakuntabhai A / Cao-Lormeau VM / Haouz A / England P / Stiasny K / Mongkolsapaya J / Heinz ...Authors: Barba-Spaeth G / Dejnirattisai W / Rouvinski A / Vaney MC / Medits I / Sharma A / Simon-Loriere E / Sakuntabhai A / Cao-Lormeau VM / Haouz A / England P / Stiasny K / Mongkolsapaya J / Heinz FX / Screaton GR / Rey FA | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30465.map.gz emd_30465.map.gz | 925.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30465-v30.xml emd-30465-v30.xml emd-30465.xml emd-30465.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
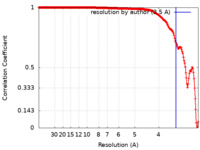
| FSC (resolution estimation) |  emd_30465_fsc.xml emd_30465_fsc.xml | 21.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_30465.png emd_30465.png | 110.9 KB | ||
| Filedesc metadata |  emd-30465.cif.gz emd-30465.cif.gz | 6.5 KB | ||
| Others |  emd_30465_additional_1.map.gz emd_30465_additional_1.map.gz emd_30465_half_map_1.map.gz emd_30465_half_map_1.map.gz emd_30465_half_map_2.map.gz emd_30465_half_map_2.map.gz | 926.5 MB 155.5 MB 155.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30465 http://ftp.pdbj.org/pub/emdb/structures/EMD-30465 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30465 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30465 | HTTPS FTP |
-Validation report
| Summary document |  emd_30465_validation.pdf.gz emd_30465_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30465_full_validation.pdf.gz emd_30465_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_30465_validation.xml.gz emd_30465_validation.xml.gz | 29.7 KB | Display | |
| Data in CIF |  emd_30465_validation.cif.gz emd_30465_validation.cif.gz | 39.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30465 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30465 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30465 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30465 | HTTPS FTP |
-Related structure data
| Related structure data |  7cthMC  7a3nC  7a3oC  7a3pC  7a3qC  7a3rC  7a3sC  7a3tC  7a3uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30465.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30465.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Dengue 2 NGC With ScFv C10 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.28 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
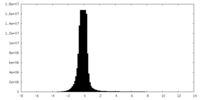
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unsharpen map
| File | emd_30465_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpen map | ||||||||||||
| Projections & Slices |
| ||||||||||||
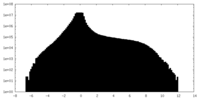
| Density Histograms |
-Half map: Half Map A
| File | emd_30465_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
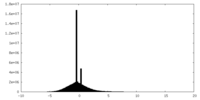
| Density Histograms |
-Half map: Half Map B
| File | emd_30465_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
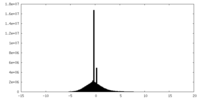
| Density Histograms |
- Sample components
Sample components
-Entire : Dengue virus
| Entire | Name:   Dengue virus Dengue virus |
|---|---|
| Components |
|
-Supramolecule #1: Dengue virus
| Supramolecule | Name: Dengue virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #2-#4 / NCBI-ID: 12637 / Sci species name: Dengue virus / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Core protein
| Macromolecule | Name: Core protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dengue virus 2 Dengue virus 2 |
| Molecular weight | Theoretical: 54.351746 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRCIGISNRD FVEGVSGGSW VDIVLEHGSC VTTMAKNKPT LDFELIKTEA KQPATLRKYC IEAKLTNTTT DSRCPTQGEP SLNEEQDKR FVCKHSMVDR GWGNGCGLFG KGGIVTCAMF TCKKNMKGKV VQPENLEYTI VITPHSGEEH AVGNDTGKHG K EIKITPQS ...String: MRCIGISNRD FVEGVSGGSW VDIVLEHGSC VTTMAKNKPT LDFELIKTEA KQPATLRKYC IEAKLTNTTT DSRCPTQGEP SLNEEQDKR FVCKHSMVDR GWGNGCGLFG KGGIVTCAMF TCKKNMKGKV VQPENLEYTI VITPHSGEEH AVGNDTGKHG K EIKITPQS SITEAELTGY GTVTMECSPR TGLDFNEMVL LQMENKAWLV HRQWFLDLPL PWLPGADTQG SNWIQKETLV TF KNPHAKK QDVVVLGSQE GAMHTALTGA TEIQMSSGNL LFTGHLKCRL RMDKLQLKGM SYSMCTGKFK VVKEIAETQH GTI VIRVQY EGDGSPCKIP FEIMDLEKRH VLGRLITVNP IVTEKDSPVN IEAEPPFGDS YIIIGVEPGQ LKLNWFKKGS SIGQ MIETT MRGAKRMAIL GDTAWDFGSL GGVFTSIGKA LHQVFGAIYG AAFSGVSWTM KILIGVIITW IGMNSRSTSL SVSLV LVGV VTLYLGVMVQ A UniProtKB: Genome polyprotein |
-Macromolecule #2: Core protein
| Macromolecule | Name: Core protein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO / EC number: flavivirin |
|---|---|
| Source (natural) | Organism:  Dengue virus 2 Dengue virus 2 |
| Molecular weight | Theoretical: 8.345762 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SVALVPHVGM GLETRTETWM SSEGAWKHAQ RIETWILRHP GFTIMAAILA YTIGTTHFQR ALIFILLTAV APSMT UniProtKB: Genome polyprotein |
-Macromolecule #3: Single Chain Variable Fragment
| Macromolecule | Name: Single Chain Variable Fragment / type: protein_or_peptide / ID: 3 Details: heavy chain variable region of the broadly neutralizing antibody EDE1 C10 Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.679234 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAEVQLVESG AEVKKPGASV KVSCKASGYT FTSYAMHWVR QAPGQRLEWM GWINAGNGNT KYSQKFQDRV TITRDTSAST AYMELSSLR SEDTAIYYCA RDKVDDYGDY WFPTLWYFDY WGQGTLVTVS SGTGGSGGGG SGGGG |
-Macromolecule #4: Single Chain Variable Fragment
| Macromolecule | Name: Single Chain Variable Fragment / type: protein_or_peptide / ID: 4 Details: light chain variable region of the broadly neutralizing antibody EDE1 C10 Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.632767 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGGGASQSAL TQPASVSGSP GQSITISCTG TSSDVGGFNY VSWFQQHPGK APKLMLYDVT SRPSGVSSRF SGSKSGNTAS LTISGLQAE DEADYYCSSH TSRGTWVFGG GTKLTVLAAA DDDDKAGWSH PQFEKGGGSG GGSGGGSWSH PQFEK |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)