[English] 日本語
 Yorodumi
Yorodumi- EMDB-3037: Density map of the engaged state of the mammalian SRP-ribosome complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3037 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Density map of the engaged state of the mammalian SRP-ribosome complex | |||||||||
 Map data Map data | This is a sharpened post-processed, masked map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosomes / SRP / mammal / translocation | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.75 Å | |||||||||
 Authors Authors | Voorhees RM / Hegde RS | |||||||||
 Citation Citation |  Journal: Elife / Year: 2015 Journal: Elife / Year: 2015Title: Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. Authors: Rebecca M Voorhees / Ramanujan S Hegde /  Abstract: The universally conserved signal recognition particle (SRP) is essential for the biogenesis of most integral membrane proteins. SRP scans the nascent chains of translating ribosomes, preferentially ...The universally conserved signal recognition particle (SRP) is essential for the biogenesis of most integral membrane proteins. SRP scans the nascent chains of translating ribosomes, preferentially engaging those with hydrophobic targeting signals, and delivers these ribosome-nascent chain complexes to the membrane. Here, we present structures of native mammalian SRP-ribosome complexes in the scanning and engaged states. These structures reveal the near-identical SRP architecture of these two states, show many of the SRP-ribosome interactions at atomic resolution, and suggest how the polypeptide-binding M domain selectively engages hydrophobic signals. The scanning M domain, pre-positioned at the ribosomal exit tunnel, is auto-inhibited by a C-terminal amphipathic helix occluding its hydrophobic binding groove. Upon engagement, the hydrophobic targeting signal displaces this amphipathic helix, which then acts as a protective lid over the signal. Biochemical experiments suggest how scanning and engagement are coordinated with translation elongation to minimize exposure of hydrophobic signals during membrane targeting. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3037.map.gz emd_3037.map.gz | 31.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3037-v30.xml emd-3037-v30.xml emd-3037.xml emd-3037.xml | 8.3 KB 8.3 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-3037.png EMD-3037.png | 325.1 KB | ||
| Others |  emd_3037_half_map_1.map.gz emd_3037_half_map_1.map.gz emd_3037_half_map_2.map.gz emd_3037_half_map_2.map.gz | 224.6 MB 224.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3037 http://ftp.pdbj.org/pub/emdb/structures/EMD-3037 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3037 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3037 | HTTPS FTP |
-Validation report
| Summary document |  emd_3037_validation.pdf.gz emd_3037_validation.pdf.gz | 340.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3037_full_validation.pdf.gz emd_3037_full_validation.pdf.gz | 340.2 KB | Display | |
| Data in XML |  emd_3037_validation.xml.gz emd_3037_validation.xml.gz | 7.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3037 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3037 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3037 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3037 | HTTPS FTP |
-Related structure data
| Related structure data |  3jajMC  3045C  3046C  3janC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3037.map.gz / Format: CCP4 / Size: 276 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3037.map.gz / Format: CCP4 / Size: 276 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a sharpened post-processed, masked map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 3037 half map 1.map
| File | emd_3037_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
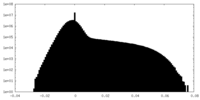
| Projections & Slices |
| ||||||||||||
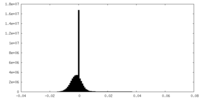
| Density Histograms |
-Supplemental map: emd 3037 half map 2.map
| File | emd_3037_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
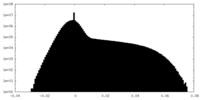
| Projections & Slices |
| ||||||||||||
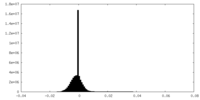
| Density Histograms |
- Sample components
Sample components
-Entire : Mammalian ribosome in complex with the signal recognition particl...
| Entire | Name: Mammalian ribosome in complex with the signal recognition particle interacting with its hydrophobic substrate |
|---|---|
| Components |
|
-Supramolecule #1000: Mammalian ribosome in complex with the signal recognition particl...
| Supramolecule | Name: Mammalian ribosome in complex with the signal recognition particle interacting with its hydrophobic substrate type: sample / ID: 1000 Oligomeric state: monomeric ribosome with monomeric signal recognition particle Number unique components: 2 |
|---|
-Supramolecule #1: mammalian ribosome
| Supramolecule | Name: mammalian ribosome / type: complex / ID: 1 / Name.synonym: 80S ribosome / Recombinant expression: No / Ribosome-details: ribosome-eukaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Signal recognition particle
| Macromolecule | Name: Signal recognition particle / type: rna / ID: 1 / Name.synonym: SRP / Details: Ribonucleoprotein particle / Classification: OTHER / Structure: OTHER / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50 mM HEPES, 200 mM KAc, 15 mM MgAc2, and 1 mM DTT |
|---|---|
| Grid | Details: Quantifoil R2/2 400 mesh copper grids. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV Method: 3uL of sampled was incubated on the grid for 30 seconds before blotting for 3 second |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Jun 16, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 27 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.75 Å / Resolution method: OTHER / Software - Name: Relion / Number images used: 52061 |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)