+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30344 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the P395-bound GPBAR-Gs complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationG protein-coupled bile acid receptor activity / adenylate cyclase-activating G protein-coupled bile acid receptor signaling pathway / positive regulation of cholangiocyte proliferation / : / cellular response to bile acid / Class A/1 (Rhodopsin-like receptors) / regulation of bicellular tight junction assembly / PKA activation in glucagon signalling / hair follicle placode formation / mu-type opioid receptor binding ...G protein-coupled bile acid receptor activity / adenylate cyclase-activating G protein-coupled bile acid receptor signaling pathway / positive regulation of cholangiocyte proliferation / : / cellular response to bile acid / Class A/1 (Rhodopsin-like receptors) / regulation of bicellular tight junction assembly / PKA activation in glucagon signalling / hair follicle placode formation / mu-type opioid receptor binding / developmental growth / corticotropin-releasing hormone receptor 1 binding / intracellular transport / D1 dopamine receptor binding / Hedgehog 'off' state / beta-2 adrenergic receptor binding / adenylate cyclase-activating adrenergic receptor signaling pathway / activation of adenylate cyclase activity / adenylate cyclase activator activity / trans-Golgi network membrane / insulin-like growth factor receptor binding / ionotropic glutamate receptor binding / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / bone development / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / cognition / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / platelet aggregation / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / adenylate cyclase-activating dopamine receptor signaling pathway / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of smell / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / GTPase binding / retina development in camera-type eye / phospholipase C-activating G protein-coupled receptor signaling pathway / Ca2+ pathway / positive regulation of cold-induced thermogenesis / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / cell population proliferation / Ras protein signal transduction / Extra-nuclear estrogen signaling / positive regulation of ERK1 and ERK2 cascade / receptor complex / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / protein-containing complex binding / GTP binding / signal transduction / extracellular exosome / membrane / metal ion binding / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Yang F / Mao C / Guo L / Lin J / Ming Q / Xiao P / Wu X / Shen Q / Guo S / Shen D ...Yang F / Mao C / Guo L / Lin J / Ming Q / Xiao P / Wu X / Shen Q / Guo S / Shen D / Lu R / Zhang L / Huang S / Ping Y / Zhang C / Ma C / Zhang K / Liang X / Shen Y / Nan F / Yi F / Luca V / Zhou J / Jiang C / Sun J / Xie X / Yu X / Zhang Y | |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structural basis of GPBAR activation and bile acid recognition. Authors: Fan Yang / Chunyou Mao / Lulu Guo / Jingyu Lin / Qianqian Ming / Peng Xiao / Xiang Wu / Qingya Shen / Shimeng Guo / Dan-Dan Shen / Ruirui Lu / Linqi Zhang / Shenming Huang / Yuqi Ping / ...Authors: Fan Yang / Chunyou Mao / Lulu Guo / Jingyu Lin / Qianqian Ming / Peng Xiao / Xiang Wu / Qingya Shen / Shimeng Guo / Dan-Dan Shen / Ruirui Lu / Linqi Zhang / Shenming Huang / Yuqi Ping / Chenlu Zhang / Cheng Ma / Kai Zhang / Xiaoying Liang / Yuemao Shen / Fajun Nan / Fan Yi / Vincent C Luca / Jiuyao Zhou / Changtao Jiang / Jin-Peng Sun / Xin Xie / Xiao Yu / Yan Zhang /   Abstract: The G-protein-coupled bile acid receptor (GPBAR) conveys the cross-membrane signalling of a vast variety of bile acids and is a signalling hub in the liver-bile acid-microbiota-metabolism axis. Here ...The G-protein-coupled bile acid receptor (GPBAR) conveys the cross-membrane signalling of a vast variety of bile acids and is a signalling hub in the liver-bile acid-microbiota-metabolism axis. Here we report the cryo-electron microscopy structures of GPBAR-G complexes stabilized by either the high-affinity P395 or the semisynthesized bile acid derivative INT-777 at 3 Å resolution. These structures revealed a large oval pocket that contains several polar groups positioned to accommodate the amphipathic cholic core of bile acids, a fingerprint of key residues to recognize diverse bile acids in the orthosteric site, a putative second bile acid-binding site with allosteric properties and structural features that contribute to bias properties. Moreover, GPBAR undertakes an atypical mode of activation and G protein coupling that features a different set of key residues connecting the ligand-binding pocket to the G-coupling site, and a specific interaction motif that is localized in intracellular loop 3. Overall, our study not only reveals unique structural features of GPBAR that are involved in bile acid recognition and allosteric effects, but also suggests the presence of distinct connecting mechanisms between the ligand-binding pocket and the G-protein-binding site in the G-protein-coupled receptor superfamily. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30344.map.gz emd_30344.map.gz | 17.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30344-v30.xml emd-30344-v30.xml emd-30344.xml emd-30344.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
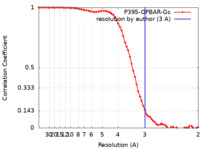
| FSC (resolution estimation) |  emd_30344_fsc.xml emd_30344_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_30344.png emd_30344.png | 74.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30344 http://ftp.pdbj.org/pub/emdb/structures/EMD-30344 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30344 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30344 | HTTPS FTP |
-Validation report
| Summary document |  emd_30344_validation.pdf.gz emd_30344_validation.pdf.gz | 396 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30344_full_validation.pdf.gz emd_30344_full_validation.pdf.gz | 395.6 KB | Display | |
| Data in XML |  emd_30344_validation.xml.gz emd_30344_validation.xml.gz | 9.5 KB | Display | |
| Data in CIF |  emd_30344_validation.cif.gz emd_30344_validation.cif.gz | 12.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30344 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30344 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30344 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30344 | HTTPS FTP |
-Related structure data
| Related structure data |  7cfmMC  7cfnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30344.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30344.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : P395-bound GPBAR-Gs complex
+Supramolecule #1: P395-bound GPBAR-Gs complex
+Supramolecule #2: GPBAR
+Supramolecule #3: P395
+Supramolecule #4: Gs
+Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
+Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
+Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
+Macromolecule #4: Nanobody-35
+Macromolecule #5: G-protein coupled bile acid receptor 1
+Macromolecule #6: 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5...
+Macromolecule #7: CHOLESTEROL
+Macromolecule #8: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 4826 / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 49310 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller