[English] 日本語
 Yorodumi
Yorodumi- EMDB-28060: Venezuelan equine encephalitis virus-like particle in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT-20, local refinement | |||||||||
 Map data Map data | Map obtained by post-processing with DeepEMhancer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Venezuelan equine encephalitis virus / neutralizing antibody / VEEV / VIRUS LIKE PARTICLE-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationtogavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host gene expression / symbiont-mediated suppression of host toll-like receptor signaling pathway / clathrin-dependent endocytosis of virus by host cell / host cell cytoplasm / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / viral envelope / host cell nucleus ...togavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host gene expression / symbiont-mediated suppression of host toll-like receptor signaling pathway / clathrin-dependent endocytosis of virus by host cell / host cell cytoplasm / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / viral envelope / host cell nucleus / virion attachment to host cell / host cell plasma membrane / structural molecule activity / virion membrane / proteolysis / RNA binding / membrane Similarity search - Function | |||||||||
| Biological species |  Venezuelan equine encephalitis virus / Venezuelan equine encephalitis virus /  | |||||||||
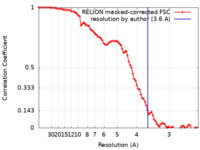
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Tsybovsky Y / Pletnev S / Verardi R / Roederer M / Kwong PD | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Vaccine elicitation and structural basis for antibody protection against alphaviruses. Authors: Matthew S Sutton / Sergei Pletnev / Victoria Callahan / Sungyoul Ko / Yaroslav Tsybovsky / Tatsiana Bylund / Ryan G Casner / Gabriele Cerutti / Christina L Gardner / Veronica Guirguis / ...Authors: Matthew S Sutton / Sergei Pletnev / Victoria Callahan / Sungyoul Ko / Yaroslav Tsybovsky / Tatsiana Bylund / Ryan G Casner / Gabriele Cerutti / Christina L Gardner / Veronica Guirguis / Raffaello Verardi / Baoshan Zhang / David Ambrozak / Margaret Beddall / Hong Lei / Eun Sung Yang / Tracy Liu / Amy R Henry / Reda Rawi / Arne Schön / Chaim A Schramm / Chen-Hsiang Shen / Wei Shi / Tyler Stephens / Yongping Yang / Maria Burgos Florez / Julie E Ledgerwood / Crystal W Burke / Lawrence Shapiro / Julie M Fox / Peter D Kwong / Mario Roederer /  Abstract: Alphaviruses are RNA viruses that represent emerging public health threats. To identify protective antibodies, we immunized macaques with a mixture of western, eastern, and Venezuelan equine ...Alphaviruses are RNA viruses that represent emerging public health threats. To identify protective antibodies, we immunized macaques with a mixture of western, eastern, and Venezuelan equine encephalitis virus-like particles (VLPs), a regimen that protects against aerosol challenge with all three viruses. Single- and triple-virus-specific antibodies were isolated, and we identified 21 unique binding groups. Cryo-EM structures revealed that broad VLP binding inversely correlated with sequence and conformational variability. One triple-specific antibody, SKT05, bound proximal to the fusion peptide and neutralized all three Env-pseudotyped encephalitic alphaviruses by using different symmetry elements for recognition across VLPs. Neutralization in other assays (e.g., chimeric Sindbis virus) yielded variable results. SKT05 bound backbone atoms of sequence-diverse residues, enabling broad recognition despite sequence variability; accordingly, SKT05 protected mice against Venezuelan equine encephalitis virus, chikungunya virus, and Ross River virus challenges. Thus, a single vaccine-elicited antibody can protect in vivo against a broad range of alphaviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28060.map.gz emd_28060.map.gz | 47.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28060-v30.xml emd-28060-v30.xml emd-28060.xml emd-28060.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28060_fsc.xml emd_28060_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_28060.png emd_28060.png | 102.8 KB | ||
| Masks |  emd_28060_msk_1.map emd_28060_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28060.cif.gz emd-28060.cif.gz | 7 KB | ||
| Others |  emd_28060_additional_1.map.gz emd_28060_additional_1.map.gz emd_28060_additional_2.map.gz emd_28060_additional_2.map.gz emd_28060_half_map_1.map.gz emd_28060_half_map_1.map.gz emd_28060_half_map_2.map.gz emd_28060_half_map_2.map.gz | 40.6 MB 5 MB 40.9 MB 41 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28060 http://ftp.pdbj.org/pub/emdb/structures/EMD-28060 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28060 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28060 | HTTPS FTP |
-Validation report
| Summary document |  emd_28060_validation.pdf.gz emd_28060_validation.pdf.gz | 973.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28060_full_validation.pdf.gz emd_28060_full_validation.pdf.gz | 972.7 KB | Display | |
| Data in XML |  emd_28060_validation.xml.gz emd_28060_validation.xml.gz | 14.2 KB | Display | |
| Data in CIF |  emd_28060_validation.cif.gz emd_28060_validation.cif.gz | 20.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28060 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28060 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28060 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28060 | HTTPS FTP |
-Related structure data
| Related structure data |  8eevMC  8decC  8dedC  8deeC  8defC  8deqC  8derC  8dulC  8dunC  8dwoC  8eeuC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28060.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28060.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map obtained by post-processing with DeepEMhancer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2315 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28060_msk_1.map emd_28060_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map before post-processing
| File | emd_28060_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map before post-processing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map obtained using standard post-processing in Relion
| File | emd_28060_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map obtained using standard post-processing in Relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1
| File | emd_28060_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2
| File | emd_28060_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between Venezuelan equine encephalitis virus-like particl...
| Entire | Name: Complex between Venezuelan equine encephalitis virus-like particle and Fab SKT20 |
|---|---|
| Components |
|
-Supramolecule #1: Complex between Venezuelan equine encephalitis virus-like particl...
| Supramolecule | Name: Complex between Venezuelan equine encephalitis virus-like particle and Fab SKT20 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The map was obtained after symmetry expansion and signal subtraction. Post-processing was performed with DeepEMhancer. |
|---|---|
| Source (natural) | Organism:  Venezuelan equine encephalitis virus Venezuelan equine encephalitis virus |
| Molecular weight | Theoretical: 33 MDa |
-Macromolecule #1: Coat protein
| Macromolecule | Name: Coat protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Venezuelan equine encephalitis virus Venezuelan equine encephalitis virus |
| Molecular weight | Theoretical: 138.662531 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFPFQPMYPM QPMPYRNPFA APRRPWFPRT DPFLAMQVQE LTRSMANLTF KQRRDAPPEG PSAKKPKKEA SQKQKGGGQG KKKKNQGKK KAKTGPPNPK AQNGNKKKTN KKPGKRQRMV MKLESDKTFP IMLEGKINGY ACVVGGKLFR PMHVEGKIDN D VLAALKTK ...String: MFPFQPMYPM QPMPYRNPFA APRRPWFPRT DPFLAMQVQE LTRSMANLTF KQRRDAPPEG PSAKKPKKEA SQKQKGGGQG KKKKNQGKK KAKTGPPNPK AQNGNKKKTN KKPGKRQRMV MKLESDKTFP IMLEGKINGY ACVVGGKLFR PMHVEGKIDN D VLAALKTK KASKYDLEYA DVPQNMRADT FKYTHEKPQG YYSWHHGAVQ YENGRFTVPK GVGAKGDSGR PILDNQGRVV AI VLGGVNE GSRTALSVVM WNEKGVTVKY TPENCEQWSL VTTMCLLANV TFPCAQPPIC YDRKPAETLA MLSVNVDNPG YDE LLEAAV KCPGRKRRST EELFNEYKLT RPYMARCIRC AVGSCHSPIA IEAVKSDGHD GYVRLQTSSQ YGLDSSGNLK GRTM RYDMH GTIKEIPLHQ VSLYTSRPCH IVDGHGYFLL ARCPAGDSIT MEFKKDSVRH SCSVPYEVKF NPVGRELYTH PPEHG VEQA CQVYAHDAQN RGAYVEMHLP GSEVDSSLVS LSGSSVTVTP PDGTSALVEC ECGGTKISET INKTKQFSQC TKKEQC RAY RLQNDKWVYN SDKLPKAAGA TLKGKLHVPF LLADGKCTVP LAPEPMITFG FRSVSLKLHP KNPTYLITRQ LADEPHY TH ELISEPAVRN FTVTEKGWEF VWGNHPPKRF WAQETAPGNP HGLPHEVITH YYHRYPMSTI LGLSICAAIA TVSVAAST W LFCRSRVACL TPYRLTPNAR IPFCLAVLCC ARTARAETTW ESLDHLWNNN QQMFWIQLLI PLAALIVVTR LLRCVCCVV PFLVMAGAAA PAYEHATTMP SQAGISYNTI VNRAGYAPLP ISITPTKIKL IPTVNLEYVT CHYKTGMDSP AIKCCGSQEC TPTYRPDEQ CKVFTGVYPF MWGGAYCFCD TENTQVSKAY VMKSDDCLAD HAEAYKAHTA SVQAFLNITV GEHSIVTTVY V NGETPVNF NGVKITAGPL STAWTPFDRK IVQYAGEIYN YDFPEYGAGQ PGAFGDIQSR TVSSSDLYAN TNLVLQRPKA GA IHVPYTQ APSGFEQWKK DKAPSLKFTA PFGCEIYTNP IRAENCAVGS IPLAFDIPDA LFTRVSETPT LSAAECTLNE CVY SSDFGG IATVKYSASK SGKCAVHVPS GTATLKEAAV ELTEQGSATI HFSTANIHPE FRLQICTSYV TCKGDCHPPK DHIV THPQY HAQTFTAAVS KTAWTWLTSL LGGSAVIIII GLVLATIVAM YVLTNQKHN UniProtKB: Structural polyprotein |
-Macromolecule #2: Fab SKT20 heavy chain
| Macromolecule | Name: Fab SKT20 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.421338 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQVQESGPG LVKPSETLSL TCAVSSGSIN DDSYYWTWIR QSPGKGLEWL GFIHGGTGKS FYNPSLESRV TISKDTSRNQ FSLTLSSVS AADTAVYYCA RSHFCSNTFC YGWFDVWGPG IRVTVSSAST KGPSVFPLAP SSRSTSESTA ALGCLVKDYF P EPVTVSWN ...String: QVQVQESGPG LVKPSETLSL TCAVSSGSIN DDSYYWTWIR QSPGKGLEWL GFIHGGTGKS FYNPSLESRV TISKDTSRNQ FSLTLSSVS AADTAVYYCA RSHFCSNTFC YGWFDVWGPG IRVTVSSAST KGPSVFPLAP SSRSTSESTA ALGCLVKDYF P EPVTVSWN SGSLTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYV CNVNHKPSNT KVDKRVEIKT CGGLEVLFQ |
-Macromolecule #3: Fab SKT20 light chain
| Macromolecule | Name: Fab SKT20 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.754445 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DVVMTQTPLS LPITPGEPAS ISCRSSQSLL HSNGNTYLHW YLQKPGQSPQ LLIYGGSNRA SGVPDRFSGS GSGTDFTLKI SKVEAEDVG VYYCVQAIAF PWTFGQGTKV EIKRTVAAPS VFIFPPSEDQ VKSGTVSVVC LLNNFYPREA SVKWKVDGAL K TGNSQESV ...String: DVVMTQTPLS LPITPGEPAS ISCRSSQSLL HSNGNTYLHW YLQKPGQSPQ LLIYGGSNRA SGVPDRFSGS GSGTDFTLKI SKVEAEDVG VYYCVQAIAF PWTFGQGTKV EIKRTVAAPS VFIFPPSEDQ VKSGTVSVVC LLNNFYPREA SVKWKVDGAL K TGNSQESV TEQDSKDNTY SLSSTLTLSS TEYQSHKVYA CEVTHQGLSS PVTKSFNRGE C |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 47.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.25 µm / Nominal defocus min: 0.75 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE |
|---|---|
| Output model |  PDB-8eev: |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)