[English] 日本語
 Yorodumi
Yorodumi- EMDB-27395: Cryo-EM local refinement of antibody SKV09 in complex with VEEV a... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM local refinement of antibody SKV09 in complex with VEEV alphavirus spike glycoprotein | |||||||||
 Map data Map data | Local Refinement after Focused Classification | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | antibody alphavirus VEEV spike trimer glycoprotein / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtogavirin / T=4 icosahedral viral capsid / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont-mediated suppression of host gene expression / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...togavirin / T=4 icosahedral viral capsid / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont-mediated suppression of host gene expression / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / RNA binding / membrane Similarity search - Function | |||||||||
| Biological species |  Venezuelan equine encephalitis virus / Venezuelan equine encephalitis virus /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.0 Å | |||||||||
 Authors Authors | Casner RG / Verardi R / Roederer M / Shapiro L | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Vaccine elicitation and structural basis for antibody protection against alphaviruses. Authors: Matthew S Sutton / Sergei Pletnev / Victoria Callahan / Sungyoul Ko / Yaroslav Tsybovsky / Tatsiana Bylund / Ryan G Casner / Gabriele Cerutti / Christina L Gardner / Veronica Guirguis / ...Authors: Matthew S Sutton / Sergei Pletnev / Victoria Callahan / Sungyoul Ko / Yaroslav Tsybovsky / Tatsiana Bylund / Ryan G Casner / Gabriele Cerutti / Christina L Gardner / Veronica Guirguis / Raffaello Verardi / Baoshan Zhang / David Ambrozak / Margaret Beddall / Hong Lei / Eun Sung Yang / Tracy Liu / Amy R Henry / Reda Rawi / Arne Schön / Chaim A Schramm / Chen-Hsiang Shen / Wei Shi / Tyler Stephens / Yongping Yang / Maria Burgos Florez / Julie E Ledgerwood / Crystal W Burke / Lawrence Shapiro / Julie M Fox / Peter D Kwong / Mario Roederer /  Abstract: Alphaviruses are RNA viruses that represent emerging public health threats. To identify protective antibodies, we immunized macaques with a mixture of western, eastern, and Venezuelan equine ...Alphaviruses are RNA viruses that represent emerging public health threats. To identify protective antibodies, we immunized macaques with a mixture of western, eastern, and Venezuelan equine encephalitis virus-like particles (VLPs), a regimen that protects against aerosol challenge with all three viruses. Single- and triple-virus-specific antibodies were isolated, and we identified 21 unique binding groups. Cryo-EM structures revealed that broad VLP binding inversely correlated with sequence and conformational variability. One triple-specific antibody, SKT05, bound proximal to the fusion peptide and neutralized all three Env-pseudotyped encephalitic alphaviruses by using different symmetry elements for recognition across VLPs. Neutralization in other assays (e.g., chimeric Sindbis virus) yielded variable results. SKT05 bound backbone atoms of sequence-diverse residues, enabling broad recognition despite sequence variability; accordingly, SKT05 protected mice against Venezuelan equine encephalitis virus, chikungunya virus, and Ross River virus challenges. Thus, a single vaccine-elicited antibody can protect in vivo against a broad range of alphaviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27395.map.gz emd_27395.map.gz | 58.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27395-v30.xml emd-27395-v30.xml emd-27395.xml emd-27395.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27395_fsc.xml emd_27395_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_27395.png emd_27395.png | 140.6 KB | ||
| Filedesc metadata |  emd-27395.cif.gz emd-27395.cif.gz | 6 KB | ||
| Others |  emd_27395_additional_1.map.gz emd_27395_additional_1.map.gz emd_27395_half_map_1.map.gz emd_27395_half_map_1.map.gz emd_27395_half_map_2.map.gz emd_27395_half_map_2.map.gz | 271.7 MB 49.5 MB 49.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27395 http://ftp.pdbj.org/pub/emdb/structures/EMD-27395 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27395 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27395 | HTTPS FTP |
-Related structure data
| Related structure data |  8deqMC  8decC  8dedC  8deeC  8defC  8derC  8dulC  8dunC  8dwoC  8eeuC  8eevC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27395.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27395.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local Refinement after Focused Classification | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.245 Å | ||||||||||||||||||||||||||||||||||||
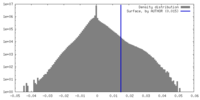
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Icosahedral VLP Map
| File | emd_27395_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Icosahedral VLP Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
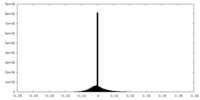
| Density Histograms |
-Half map: half map 1
| File | emd_27395_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_27395_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM local refinement of antibody SKV09 in complex with VEEV a...
| Entire | Name: Cryo-EM local refinement of antibody SKV09 in complex with VEEV alphavirus spike glycoprotein |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM local refinement of antibody SKV09 in complex with VEEV a...
| Supramolecule | Name: Cryo-EM local refinement of antibody SKV09 in complex with VEEV alphavirus spike glycoprotein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Venezuelan equine encephalitis virus Venezuelan equine encephalitis virus |
-Macromolecule #1: Spike glycoprotein E1
| Macromolecule | Name: Spike glycoprotein E1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Venezuelan equine encephalitis virus Venezuelan equine encephalitis virus |
| Molecular weight | Theoretical: 43.668984 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YEHATTMPSQ AGISYNTIVN RAGYAPLPIS ITPTKIKLIP TVNLEYVTCH YKTGMDSPAI KCCGSQECTP TYRPDEQCKV FTGVYPFMW GGAYCFCDTE NTQVSKAYVM KSDDCLADHA EAYKAHTASV QAFLNITVGE HSIVTTVYVN GETPVNFNGV K ITAGPLST ...String: YEHATTMPSQ AGISYNTIVN RAGYAPLPIS ITPTKIKLIP TVNLEYVTCH YKTGMDSPAI KCCGSQECTP TYRPDEQCKV FTGVYPFMW GGAYCFCDTE NTQVSKAYVM KSDDCLADHA EAYKAHTASV QAFLNITVGE HSIVTTVYVN GETPVNFNGV K ITAGPLST AWTPFDRKIV QYAGEIYNYD FPEYGAGQPG AFGDIQSRTV SSSDLYANTN LVLQRPKAGA IHVPYTQAPS GF EQWKKDK APSLKFTAPF GCEIYTNPIR AENCAVGSIP LAFDIPDALF TRVSETPTLS AAECTLNECV YSSDFGGIAT VKY SASKSG KCAVHVPSGT ATLKEAAVEL TEQGSATIHF STANIHPEFR LQICTSYVTC KGDCHPPKDH IVTHPQYHAQ TFTA AV UniProtKB: Structural polyprotein |
-Macromolecule #2: SKV09 Fab Heavy Chain
| Macromolecule | Name: SKV09 Fab Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.095468 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGPG LVKPSETLSL TCTVSGGSIS GYYWNWIRQS PGKGLEWIGN MYDSTGDTNY NPSLKSRVTL SIDTSKNQFS MKLNSVTAA DTAVYFCASG SGDWFGYFYR WGQGVLVTVS S |
-Macromolecule #3: SKV09 Fab Light Chain
| Macromolecule | Name: SKV09 Fab Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.761066 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDTVT ITCRASQSIS SWLAWYQQKP GKAPNLLIYK ASSLQTGVPS RFSGSGSETH FTLTISSLQS EDFATYYCQ QYISRPWTFG QGTKVEIK |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)