+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Overall map of Arabidopsis SPY in complex with GDP-fucose | |||||||||
 Map data Map data | NU refinement Overall Map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
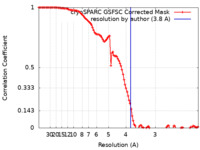
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Kumar S / Zhou Y / Lucas D / Borgnia MJ / Bartesaghi A / Zhou P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs Authors: Kumar S / Zhou Y | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27700.map.gz emd_27700.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27700-v30.xml emd-27700-v30.xml emd-27700.xml emd-27700.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27700_fsc.xml emd_27700_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_27700.png emd_27700.png | 68.6 KB | ||
| Masks |  emd_27700_msk_1.map emd_27700_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_27700_half_map_1.map.gz emd_27700_half_map_1.map.gz emd_27700_half_map_2.map.gz emd_27700_half_map_2.map.gz | 115.7 MB 115.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27700 http://ftp.pdbj.org/pub/emdb/structures/EMD-27700 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27700 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27700 | HTTPS FTP |
-Validation report
| Summary document |  emd_27700_validation.pdf.gz emd_27700_validation.pdf.gz | 653.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27700_full_validation.pdf.gz emd_27700_full_validation.pdf.gz | 653.1 KB | Display | |
| Data in XML |  emd_27700_validation.xml.gz emd_27700_validation.xml.gz | 19 KB | Display | |
| Data in CIF |  emd_27700_validation.cif.gz emd_27700_validation.cif.gz | 24.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27700 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27700 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27700 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27700 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27700.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27700.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | NU refinement Overall Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
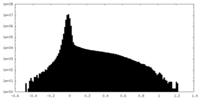
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27700_msk_1.map emd_27700_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
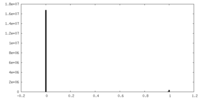
| Density Histograms |
-Half map: Half map 1
| File | emd_27700_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
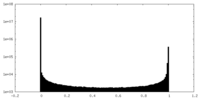
| Density Histograms |
-Half map: Half map 2
| File | emd_27700_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
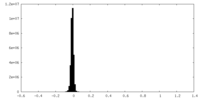
| Density Histograms |
- Sample components
Sample components
-Entire : SPY GDP-fucose complex
| Entire | Name: SPY GDP-fucose complex |
|---|---|
| Components |
|
-Supramolecule #1: SPY GDP-fucose complex
| Supramolecule | Name: SPY GDP-fucose complex / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: SPY GDP-fucose complex
| Macromolecule | Name: SPY GDP-fucose complex / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVGLEDDTER ERSPVVENGF SNGSRSSSSS AGVLSPSRKV TQGNDTLSYA NILRARNKFA DALALYEAML EKDSKNVEAH IGKGICLQTQ NKGNLAFDCF SEAIRLDPHN ACALTHCGIL HKEEGRLVEA AESYQKALMA DASYKPAAEC LAIVLTDLGT SLKLAGNTQE ...String: MVGLEDDTER ERSPVVENGF SNGSRSSSSS AGVLSPSRKV TQGNDTLSYA NILRARNKFA DALALYEAML EKDSKNVEAH IGKGICLQTQ NKGNLAFDCF SEAIRLDPHN ACALTHCGIL HKEEGRLVEA AESYQKALMA DASYKPAAEC LAIVLTDLGT SLKLAGNTQE GIQKYYEALK IDPHYAPAYY NLGVVYSEMM QYDNALSCYE KAALERPMYA EAYCNMGVIY KNRGDLEMAI TCYERCLAVS PNFEIAKNNM AIALTDLGTK VKLEGDVTQG VAYYKKALYY NWHYADAMYN LGVAYGEMLK FDMAIVFYEL AFHFNPHCAE ACNNLGVLYK DRDNLDKAVE CYQMALSIKP NFAQSLNNLG VVYTVQGKMD AAASMIEKAI LANPTYAEAF NNLGVLYRDA GNITMAIDAY EECLKIDPDS RNAGQNRLLA MNYINEGLDD KLFEAHRDWG WRFTRLHPQY TSWDNLKDPE RPITIGYISP DFFTHSVSYF IEAPLTHHDY TKYKVVVYSA VVKADAKTYR FRDKVLKKGG VWKDIYGIDE KKIASMVRED KIDILVELTG HTANNKLGTM ACRPAPVQVT WIGYPNTTGL PTVDYRITDS LADPPDTKQK QVEELVRLPD CFLCYTPSPE AGPVCPTPAL SNGFVTFGSF NNLAKITPKV LQVWARILCA VPNSRLVVKC KPFCCDSIRQ RFLTTLEQLG LESKRVDLLP LILFNHDHMQ AYSLMDISLD TFPYAGTTTT CESLYMGVPC VTMAGSVHAH NVGVSLLTKV GLGHLVAKNE DEYVQLSVDL ASDVTALSKL RMSLRDLMAG SPVCNGPSFA VGLESAYRNM WKKYCKGEVP SLRRMEMLQK EVHDDPLISK DLGPSRVSVT GEATPSLKAN GSAPVPSSLP TQSPQLSKRM DSTSGGSENL YFQGGSHHHH HHHHHHGGWS HPQFEK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 40.0 nm / Pretreatment - Type: PLASMA CLEANING | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 289.15 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Average electron dose: 52.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)