[English] 日本語
 Yorodumi
Yorodumi- EMDB-27702: Focused map (monomer B) for Arabidopsis SPY in complex with GDP-fucose -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Focused map (monomer B) for Arabidopsis SPY in complex with GDP-fucose | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | O-Fucosyltransferase / PLANT PROTEIN | |||||||||
| Biological species |  | |||||||||
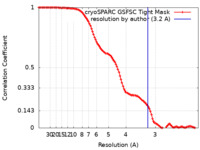
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Kumar S / Zhou Y / Lucas D / Borgnia MJ / Bartesaghi A / Zhou P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure and dynamics of the Arabidopsis O-fucosyltransferase SPINDLY. Authors: Shivesh Kumar / Yan Wang / Ye Zhou / Lucas Dillard / Fay-Wei Li / Carly A Sciandra / Ning Sui / Rodolfo Zentella / Emily Zahn / Jeffrey Shabanowitz / Donald F Hunt / Mario J Borgnia / ...Authors: Shivesh Kumar / Yan Wang / Ye Zhou / Lucas Dillard / Fay-Wei Li / Carly A Sciandra / Ning Sui / Rodolfo Zentella / Emily Zahn / Jeffrey Shabanowitz / Donald F Hunt / Mario J Borgnia / Alberto Bartesaghi / Tai-Ping Sun / Pei Zhou /  Abstract: SPINDLY (SPY) in Arabidopsis thaliana is a novel nucleocytoplasmic protein O-fucosyltransferase (POFUT), which regulates diverse developmental processes. Sequence analysis indicates that SPY is ...SPINDLY (SPY) in Arabidopsis thaliana is a novel nucleocytoplasmic protein O-fucosyltransferase (POFUT), which regulates diverse developmental processes. Sequence analysis indicates that SPY is distinct from ER-localized POFUTs and contains N-terminal tetratricopeptide repeats (TPRs) and a C-terminal catalytic domain resembling the O-linked-N-acetylglucosamine (GlcNAc) transferases (OGTs). However, the structural feature that determines the distinct enzymatic selectivity of SPY remains unknown. Here we report the cryo-electron microscopy (cryo-EM) structure of SPY and its complex with GDP-fucose, revealing distinct active-site features enabling GDP-fucose instead of UDP-GlcNAc binding. SPY forms an antiparallel dimer instead of the X-shaped dimer in human OGT, and its catalytic domain interconverts among multiple conformations. Analysis of mass spectrometry, co-IP, fucosylation activity, and cryo-EM data further demonstrates that the N-terminal disordered peptide in SPY contains trans auto-fucosylation sites and inhibits the POFUT activity, whereas TPRs 1-5 dynamically regulate SPY activity by interfering with protein substrate binding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27702.map.gz emd_27702.map.gz | 118.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27702-v30.xml emd-27702-v30.xml emd-27702.xml emd-27702.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27702_fsc.xml emd_27702_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_27702.png emd_27702.png | 47.2 KB | ||
| Masks |  emd_27702_msk_1.map emd_27702_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_27702_half_map_1.map.gz emd_27702_half_map_1.map.gz emd_27702_half_map_2.map.gz emd_27702_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27702 http://ftp.pdbj.org/pub/emdb/structures/EMD-27702 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27702 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27702 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27702.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27702.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27702_msk_1.map emd_27702_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_27702_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_27702_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SPY GPD-complex
| Entire | Name: SPY GPD-complex |
|---|---|
| Components |
|
-Supramolecule #1: SPY GPD-complex
| Supramolecule | Name: SPY GPD-complex / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 40 / Pretreatment - Type: PLASMA CLEANING | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 289.15 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Average electron dose: 52.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)