[English] 日本語
 Yorodumi
Yorodumi- EMDB-27347: nsEM map of hemagglutinin H1 A/Mich/045/15 complexed with polyclo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
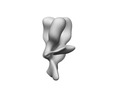
| Title | nsEM map of hemagglutinin H1 A/Mich/045/15 complexed with polyclonal Fab samples from individual 182419 at day 0 using Sulfo-SDAD photo-cross-linker | ||||||||||||
 Map data Map data | nsEM map of hemagglutinin H1 A/Mich/045/15 complexed with polyclonal Fab samples from individual 182419 at day 0 using Sulfo-SDAD photo-cross-linker | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | influenza / H1 / nsEM / VIRAL PROTEIN | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.0 Å | ||||||||||||
 Authors Authors | Torrents de la Pena A / Sewall LM / Ward AB | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Rep Methods / Year: 2023 Journal: Cell Rep Methods / Year: 2023Title: Increasing sensitivity of antibody-antigen interactions using photo-cross-linking. Authors: Alba Torrents de la Peña / Leigh M Sewall / Rebeca de Paiva Froes Rocha / Abigail M Jackson / Payal P Pratap / Sandhya Bangaru / Christopher A Cottrell / Subhasis Mohanty / Albert C Shaw / Andrew B Ward /  Abstract: Understanding antibody-antigen interactions in a polyclonal immune response in humans and animal models is critical for rational vaccine design. Current approaches typically characterize antibodies ...Understanding antibody-antigen interactions in a polyclonal immune response in humans and animal models is critical for rational vaccine design. Current approaches typically characterize antibodies that are functionally relevant or highly abundant. Here, we use photo-cross-linking and single-particle electron microscopy to increase antibody detection and unveil epitopes of low-affinity and low-abundance antibodies, leading to a broader structural characterization of polyclonal immune responses. We employed this approach across three different viral glycoproteins and showed increased sensitivity of detection relative to currently used methods. Results were most noticeable in early and late time points of a polyclonal immune response. Additionally, the use of photo-cross-linking revealed intermediate antibody binding states and demonstrated a distinctive way to study antibody binding mechanisms. This technique can be used to structurally characterize the landscape of a polyclonal immune response of patients in vaccination or post-infection studies at early time points, allowing for rapid iterative design of vaccine immunogens. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27347.map.gz emd_27347.map.gz | 23.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27347-v30.xml emd-27347-v30.xml emd-27347.xml emd-27347.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27347.png emd_27347.png | 18.8 KB | ||
| Others |  emd_27347_half_map_1.map.gz emd_27347_half_map_1.map.gz emd_27347_half_map_2.map.gz emd_27347_half_map_2.map.gz | 23.5 MB 23.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27347 http://ftp.pdbj.org/pub/emdb/structures/EMD-27347 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27347 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27347 | HTTPS FTP |
-Validation report
| Summary document |  emd_27347_validation.pdf.gz emd_27347_validation.pdf.gz | 570.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27347_full_validation.pdf.gz emd_27347_full_validation.pdf.gz | 570.4 KB | Display | |
| Data in XML |  emd_27347_validation.xml.gz emd_27347_validation.xml.gz | 10.8 KB | Display | |
| Data in CIF |  emd_27347_validation.cif.gz emd_27347_validation.cif.gz | 12.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27347 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27347 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27347 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27347 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27347.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27347.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | nsEM map of hemagglutinin H1 A/Mich/045/15 complexed with polyclonal Fab samples from individual 182419 at day 0 using Sulfo-SDAD photo-cross-linker | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.06 Å | ||||||||||||||||||||||||||||||||||||
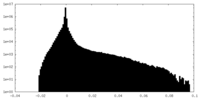
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: nsEM half map of hemagglutinin H1 A/Mich/045/15 complexed...
| File | emd_27347_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | nsEM half map of hemagglutinin H1 A/Mich/045/15 complexed with polyclonal Fab samples from individual 182419 at day 0 using Sulfo-SDAD photo-cross-linker | ||||||||||||
| Projections & Slices |
| ||||||||||||
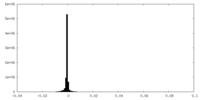
| Density Histograms |
-Half map: nsEM half map of hemagglutinin H1 A/Mich/045/15 complexed...
| File | emd_27347_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | nsEM half map of hemagglutinin H1 A/Mich/045/15 complexed with polyclonal Fab samples from individual 182419 at day 0 using Sulfo-SDAD photo-cross-linker | ||||||||||||
| Projections & Slices |
| ||||||||||||
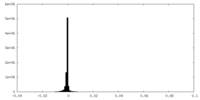
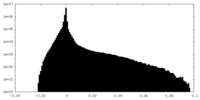
| Density Histograms |
- Sample components
Sample components
-Entire : nsEM map of hemagglutinin H1 A/Mich/045/15 complexed with polyclo...
| Entire | Name: nsEM map of hemagglutinin H1 A/Mich/045/15 complexed with polyclonal Fab samples from individual 182419 at day 0 using Sulfo-SDAD photo-cross-linker |
|---|---|
| Components |
|
-Supramolecule #1: nsEM map of hemagglutinin H1 A/Mich/045/15 complexed with polyclo...
| Supramolecule | Name: nsEM map of hemagglutinin H1 A/Mich/045/15 complexed with polyclonal Fab samples from individual 182419 at day 0 using Sulfo-SDAD photo-cross-linker type: complex / ID: 1 / Parent: 0 Details: HA recombinantly expressed in HEK 293F cells Fab samples isolated from several individuals at different timepoints |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Concentration: 0.02 0.020 / Component - Formula: TBS / Component - Name: Tris-buffered saline |
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
| Details | 15 ug of H1 A/Mich15/045/15 was complexed overnight with 500 ug of polyclonal Fab samples and photo-cross-linked. Sample was SEC purified and the complex peak was concentrated to ~0.02 mg/ml and imaged |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Number grids imaged: 1 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 52000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)