+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | gRAMP non-matching PFS-with Mg | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / GRAMP / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology | : / CRISPR type III-associated protein / RAMP superfamily / defense response to virus / RNA binding / RAMP superfamily protein Function and homology information Function and homology information | |||||||||
| Biological species |  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.62 Å | |||||||||
 Authors Authors | Hu C / Nam KH / Schuler G / Ke A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Craspase is a CRISPR RNA-guided, RNA-activated protease. Authors: Chunyi Hu / Sam P B van Beljouw / Ki Hyun Nam / Gabriel Schuler / Fran Ding / Yanru Cui / Alicia Rodríguez-Molina / Anna C Haagsma / Menno Valk / Martin Pabst / Stan J J Brouns / Ailong Ke /    Abstract: The CRISPR-Cas type III-E RNA-targeting effector complex gRAMP/Cas7-11 is associated with a caspase-like protein (TPR-CHAT/Csx29) to form Craspase (CRISPR-guided caspase). Here, we use cryo-electron ...The CRISPR-Cas type III-E RNA-targeting effector complex gRAMP/Cas7-11 is associated with a caspase-like protein (TPR-CHAT/Csx29) to form Craspase (CRISPR-guided caspase). Here, we use cryo-electron microscopy snapshots of Craspase to explain its target RNA cleavage and protease activation mechanisms. Target-guide pairing extending into the 5' region of the guide RNA displaces a gating loop in gRAMP, which triggers an extensive conformational relay that allosterically aligns the protease catalytic dyad and opens an amino acid side-chain-binding pocket. We further define Csx30 as the endogenous protein substrate that is site-specifically proteolyzed by RNA-activated Craspase. This protease activity is switched off by target RNA cleavage by gRAMP and is not activated by RNA targets containing a matching protospacer flanking sequence. We thus conclude that Craspase is a target RNA-activated protease with self-regulatory capacity. | |||||||||
| History |
|
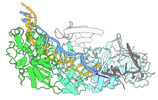
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27263.map.gz emd_27263.map.gz | 915.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27263-v30.xml emd-27263-v30.xml emd-27263.xml emd-27263.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27263.png emd_27263.png | 96.8 KB | ||
| Filedesc metadata |  emd-27263.cif.gz emd-27263.cif.gz | 6.3 KB | ||
| Others |  emd_27263_half_map_1.map.gz emd_27263_half_map_1.map.gz emd_27263_half_map_2.map.gz emd_27263_half_map_2.map.gz | 77.8 MB 77.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27263 http://ftp.pdbj.org/pub/emdb/structures/EMD-27263 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27263 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27263 | HTTPS FTP |
-Validation report
| Summary document |  emd_27263_validation.pdf.gz emd_27263_validation.pdf.gz | 1002.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27263_full_validation.pdf.gz emd_27263_full_validation.pdf.gz | 1001.7 KB | Display | |
| Data in XML |  emd_27263_validation.xml.gz emd_27263_validation.xml.gz | 13.1 KB | Display | |
| Data in CIF |  emd_27263_validation.cif.gz emd_27263_validation.cif.gz | 14.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27263 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27263 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27263 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27263 | HTTPS FTP |
-Related structure data
| Related structure data |  8d9iMC  8d8nC  8d97C  8d9eC  8d9fC  8d9gC  8d9hC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27263.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27263.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.284 Å | ||||||||||||||||||||||||||||||||||||
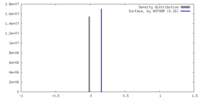
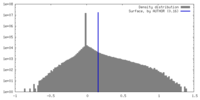
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_27263_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
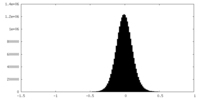
| Density Histograms |
-Half map: #1
| File | emd_27263_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
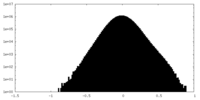
| Density Histograms |
- Sample components
Sample components
-Entire : gRAMP-non matching PFS-with Mg
| Entire | Name: gRAMP-non matching PFS-with Mg |
|---|---|
| Components |
|
-Supramolecule #1: gRAMP-non matching PFS-with Mg
| Supramolecule | Name: gRAMP-non matching PFS-with Mg / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
-Macromolecule #1: RAMP superfamily protein
| Macromolecule | Name: RAMP superfamily protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
| Molecular weight | Theoretical: 143.1015 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNITVELTFF EPYRLVEWFD WDARKKSHSA MRGQAFAQWT WKGKGRTAGK SFITGTLVRS AVIKAVEELL SLNNGKWEGV PCCNGSFQT DESKGKKPSF LRKRHTLQWQ ANNKNICDKE EACPFCILLG RFDNAGKVHE RNKDYDIHFS NFDLDHKQEK N DLRLVDIA ...String: MNITVELTFF EPYRLVEWFD WDARKKSHSA MRGQAFAQWT WKGKGRTAGK SFITGTLVRS AVIKAVEELL SLNNGKWEGV PCCNGSFQT DESKGKKPSF LRKRHTLQWQ ANNKNICDKE EACPFCILLG RFDNAGKVHE RNKDYDIHFS NFDLDHKQEK N DLRLVDIA SGRILNRVDF DTGKAKDYFR TWEADYETYG TYTGRITLRN EHAKKLLLAS LGFVDKLCGA LCRIEVIKKE VL SEDHNDE LRKQAEVIVE AFKQNDKLEK IRILADAIRT LRLHGEGVIE KDELPDGKEE RDKGHHLWDI KVQGTALRTK LKE LWQSNK DIGWRKFTEM LGSNLYLIYK KETGIETKEW IIVGRLKAAT PFYFGVQQPS DSIPGVINEH TSFNILLDKE NRYR IPRSA LRGALRRDLR TAFGSGCNVS LGGQILCNCK VCIEMRRITL KDSVSDFSEP PEIRYRIAKN PGTATVEDGS LFDIE VGPE GLTFPFVLRY RGHKFPEQLS SVIRYWEEND GKNGMAWLGG LDSTGKGRFA LKDIKIFEWD LNQKINEYIK ERGMRG KEK ELLEMGESSL PDGLIPYKFF EERECLFPYK ENLKPQWSEV QYTIEVGSPL LTADTISALT EPGNRDAIAY KKRVYND GN NAIEPEPRFA VKSETHRGIF RTAVGRRTGD LGKEDHEDCT CDMCIIFGNE HESSKIRFED LELINGNEFE KLEKHIDH V AIDRFTGGAL DKAKFDTYPL AGSPKKPLKL KGRFWIKKGF SGDHKLLITT ALSDIRDGLY PLGSKGGVGY GWVAGISID DNVPDDFKEM INKTEAAAAA AAAAAAAAAA AAAAKNKNIY YPHYFLDSGS KVYREKDIIT HEEFTEELLS GKINCKLETL TPLIIPDTS DENGLKLQGN KPGHKNYKFF NINGELMIPG SELRGMLRTH FEALTKSCFA IFGEDSTLSW RRKCASKTLG G KLDKALHP CTGLSDGLCP GCHLFGTTDY KGRVKFGFAK YENGPEWLIT RGNNPERSLT LGVLESPRPA FSIPDDESEI PG RKFYLHH NGWRIIRQKQ LEIRETVQPE RNVTTEVMDK GNVFSFDVKF ENLREWELGL LLQSLDPGKN IAHKLGKGKP YGF GSVKIK IDSLHTFKIN SNNDKIKRVP QSDIREYINK GYQKLIEWSG NNSIQKGNVL PQWHVIPHID KLYKLLWVPF LNDS KLEPD VRYPVLNEES KGYIEGSDYT YKKLGDKDNL PYKTRVKGLT TPWSPWNPFQ V UniProtKB: RAMP superfamily protein, RAMP superfamily protein |
-Macromolecule #2: RNA (5'-R(P*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP...
| Macromolecule | Name: RNA (5'-R(P*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*A)-3') type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
| Molecular weight | Theoretical: 6.180762 KDa |
| Sequence | String: UCCGGGGCAG AAAAUUGGA |
-Macromolecule #3: RNA (35-MER)
| Macromolecule | Name: RNA (35-MER) / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
| Molecular weight | Theoretical: 11.078582 KDa |
| Sequence | String: ACUUAAUGUC ACGGUACCCA AUUUUCUGCC CCGGA |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.62 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 79785 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)