+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of WAVE Regulatory Complex | |||||||||
 マップデータ マップデータ | WRC230VCA complex sharpened map | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | actin regulator / GTPase binding protein / cytoskeletal regulator / CELL INVASION | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報peripheral region of growth cone / SCAR complex / negative regulation of synaptic vesicle recycling / modification of synaptic structure / positive regulation of neurotrophin TRK receptor signaling pathway / lamellipodium morphogenesis / positive regulation of Arp2/3 complex-mediated actin nucleation / Arp2/3 complex binding / regulation of actin polymerization or depolymerization / central region of growth cone ...peripheral region of growth cone / SCAR complex / negative regulation of synaptic vesicle recycling / modification of synaptic structure / positive regulation of neurotrophin TRK receptor signaling pathway / lamellipodium morphogenesis / positive regulation of Arp2/3 complex-mediated actin nucleation / Arp2/3 complex binding / regulation of actin polymerization or depolymerization / central region of growth cone / modification of postsynaptic actin cytoskeleton / dendrite extension / regulation of translation at postsynapse, modulating synaptic transmission / filopodium tip / regulation of actin filament polymerization / regulation of modification of postsynaptic actin cytoskeleton / RNA 7-methylguanosine cap binding / ruffle organization / cell projection assembly / axon extension / positive regulation of ruffle assembly / cortical actin cytoskeleton organization / lamellipodium assembly / regulation of myelination / positive regulation of dendrite development / protein kinase A binding / positive regulation of actin filament polymerization / protein kinase A regulatory subunit binding / Rac protein signal transduction / filamentous actin / dendritic growth cone / lamellipodium membrane / excitatory synapse / RHOG GTPase cycle / RHO GTPases Activate WASPs and WAVEs / RAC2 GTPase cycle / RAC3 GTPase cycle / positive regulation of axon extension / response to electrical stimulus / axonal growth cone / positive regulation of lamellipodium assembly / ruffle / cellular response to brain-derived neurotrophic factor stimulus / translation repressor activity / RAC1 GTPase cycle / actin filament polymerization / receptor-mediated endocytosis / neuron projection morphogenesis / axon guidance / actin filament organization / central nervous system development / filopodium / mitochondrion organization / FCGR3A-mediated phagocytosis / cell motility / positive regulation of protein-containing complex assembly / terminal bouton / cell morphogenesis / small GTPase binding / Regulation of actin dynamics for phagocytic cup formation / cognition / VEGFA-VEGFR2 Pathway / cellular response to insulin stimulus / specific granule lumen / positive regulation of fibroblast proliferation / actin filament binding / cell migration / actin cytoskeleton / tertiary granule lumen / lamellipodium / regulation of translation / regulation of cell shape / actin binding / actin cytoskeleton organization / protein-containing complex assembly / fibroblast proliferation / secretory granule lumen / in utero embryonic development / dendritic spine / mitochondrial outer membrane / cytoskeleton / postsynapse / neuron projection / intracellular membrane-bounded organelle / focal adhesion / neuronal cell body / apoptotic process / synapse / Neutrophil degranulation / protein-containing complex binding / perinuclear region of cytoplasm / protein-containing complex / extracellular exosome / extracellular region / nucleoplasm / identical protein binding / cytosol 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
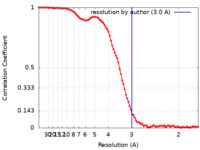
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.0 Å | |||||||||
 データ登録者 データ登録者 | Ding B / Yang S / Chen B / Chowdhury S | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2022 ジャーナル: Nat Commun / 年: 2022タイトル: Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase. 著者: Bojian Ding / Sheng Yang / Matthias Schaks / Yijun Liu / Abbigale J Brown / Klemens Rottner / Saikat Chowdhury / Baoyu Chen /    要旨: The Rho-family GTPase Rac1 activates the WAVE regulatory complex (WRC) to drive Arp2/3 complex-mediated actin polymerization in many essential processes. Rac1 binds to WRC at two distinct sites-the ...The Rho-family GTPase Rac1 activates the WAVE regulatory complex (WRC) to drive Arp2/3 complex-mediated actin polymerization in many essential processes. Rac1 binds to WRC at two distinct sites-the A and D sites. Precisely how Rac1 binds and how the binding triggers WRC activation remain unknown. Here we report WRC structures by itself, and when bound to single or double Rac1 molecules, at ~3 Å resolutions by cryogenic-electron microscopy. The structures reveal that Rac1 binds to the two sites by distinct mechanisms, and binding to the A site, but not the D site, drives WRC activation. Activation involves a series of unique conformational changes leading to the release of sequestered WCA (WH2-central-acidic) polypeptide, which stimulates the Arp2/3 complex to polymerize actin. Together with biochemical and cellular analyses, the structures provide a novel mechanistic understanding of how the Rac1-WRC-Arp2/3-actin signaling axis is regulated in diverse biological processes and diseases. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_26732.map.gz emd_26732.map.gz | 8.4 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-26732-v30.xml emd-26732-v30.xml emd-26732.xml emd-26732.xml | 28.5 KB 28.5 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_26732_fsc.xml emd_26732_fsc.xml | 11.7 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_26732.png emd_26732.png | 119.8 KB | ||
| マスクデータ |  emd_26732_msk_1.map emd_26732_msk_1.map | 64 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-26732.cif.gz emd-26732.cif.gz | 8.7 KB | ||
| その他 |  emd_26732_additional_1.map.gz emd_26732_additional_1.map.gz emd_26732_half_map_1.map.gz emd_26732_half_map_1.map.gz emd_26732_half_map_2.map.gz emd_26732_half_map_2.map.gz | 8.4 MB 57.1 MB 57.1 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26732 http://ftp.pdbj.org/pub/emdb/structures/EMD-26732 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26732 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26732 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_26732_validation.pdf.gz emd_26732_validation.pdf.gz | 895.1 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_26732_full_validation.pdf.gz emd_26732_full_validation.pdf.gz | 894.7 KB | 表示 | |
| XML形式データ |  emd_26732_validation.xml.gz emd_26732_validation.xml.gz | 16 KB | 表示 | |
| CIF形式データ |  emd_26732_validation.cif.gz emd_26732_validation.cif.gz | 21.1 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26732 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26732 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26732 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26732 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_26732.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_26732.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | WRC230VCA complex sharpened map | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.8757 Å | ||||||||||||||||||||||||||||||||||||
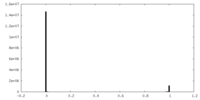
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_26732_msk_1.map emd_26732_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
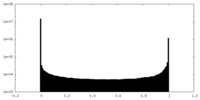
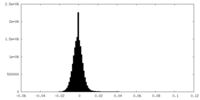
| 投影像・断面図 |
| ||||||||||||
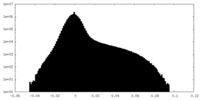
| 密度ヒストグラム |
-追加マップ: WRC230VCA complex masked unsharpened map
| ファイル | emd_26732_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | WRC230VCA complex masked unsharpened map | ||||||||||||
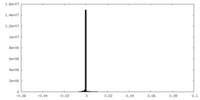
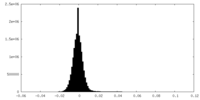
| 投影像・断面図 |
| ||||||||||||
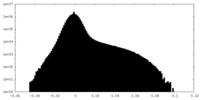
| 密度ヒストグラム |
-ハーフマップ: WRC230VCA complex unfiltered unmasked unsharpened first half map
| ファイル | emd_26732_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | WRC230VCA complex unfiltered unmasked unsharpened first half map | ||||||||||||
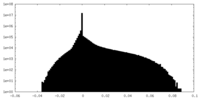
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: WRC230VCA complex unfiltered unmasked unsharpened second half map
| ファイル | emd_26732_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | WRC230VCA complex unfiltered unmasked unsharpened second half map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : WAVE regulatory complex
| 全体 | 名称: WAVE regulatory complex |
|---|---|
| 要素 |
|
-超分子 #1: WAVE regulatory complex
| 超分子 | 名称: WAVE regulatory complex / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 340 KDa |
-分子 #1: Cytoplasmic FMR1-interacting protein 1
| 分子 | 名称: Cytoplasmic FMR1-interacting protein 1 / タイプ: protein_or_peptide / ID: 1 詳細: This construct contains two additional uncleaved residues "GA" in the N terminus from the construct design and purification procedure. Densities for these residues are not observed in the map ...詳細: This construct contains two additional uncleaved residues "GA" in the N terminus from the construct design and purification procedure. Densities for these residues are not observed in the map and were not included in the sample sequence to avoid numbering shifts. コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 145.36375 KDa |
| 組換発現 | 生物種:  Trichoplusia ni (イラクサキンウワバ) Trichoplusia ni (イラクサキンウワバ) |
| 配列 | 文字列: MAAQVTLEDA LSNVDLLEEL PLPDQQPCIE PPPSSLLYQP NFNTNFEDRN AFVTGIARYI EQATVHSSMN EMLEEGQEYA VMLYTWRSC SRAIPQVKCN EQPNRVEIYE KTVEVLEPEV TKLMNFMYFQ RNAIERFCGE VRRLCHAERR KDFVSEAYLI T LGKFINMF ...文字列: MAAQVTLEDA LSNVDLLEEL PLPDQQPCIE PPPSSLLYQP NFNTNFEDRN AFVTGIARYI EQATVHSSMN EMLEEGQEYA VMLYTWRSC SRAIPQVKCN EQPNRVEIYE KTVEVLEPEV TKLMNFMYFQ RNAIERFCGE VRRLCHAERR KDFVSEAYLI T LGKFINMF AVLDELKNMK CSVKNDHSAY KRAAQFLRKM ADPQSIQESQ NLSMFLANHN KITQSLQQQL EVISGYEELL AD IVNLCVD YYENRMYLTP SEKHMLLKVM GFGLYLMDGS VSNIYKLDAK KRINLSKIDK YFKQLQVVPL FGDMQIELAR YIK TSAHYE ENKSRWTCTS SGSSPQYNIC EQMIQIREDH MRFISELARY SNSEVVTGSG RQEAQKTDAE YRKLFDLALQ GLQL LSQWS AHVMEVYSWK LVHPTDKYSN KDCPDSAEEY ERATRYNYTS EEKFALVEVI AMIKGLQVLM GRMESVFNHA IRHTV YAAL QDFSQVTLRE PLRQAIKKKK NVIQSVLQAI RKTVCDWETG HEPFNDPALR GEKDPKSGFD IKVPRRAVGP SSTQLY MVR TMLESLIADK SGSKKTLRSS LEGPTILDIE KFHRESFFYT HLINFSETLQ QCCDLSQLWF REFFLELTMG RRIQFPI EM SMPWILTDHI LETKEASMME YVLYSLDLYN DSAHYALTRF NKQFLYDEIE AEVNLCFDQF VYKLADQIFA YYKVMAGS L LLDKRLRSEC KNQGATIHLP PSNRYETLLK QRHVQLLGRS IDLNRLITQR VSAAMYKSLE LAIGRFESED LTSIVELDG LLEINRMTHK LLSRYLTLDG FDAMFREANH NVSAPYGRIT LHVFWELNYD FLPNYCYNGS TNRFVRTVLP FSQEFQRDKQ PNAQPQYLH GSKALNLAYS SIYGSYRNFV GPPHFQVICR LLGYQGIAVV MEELLKVVKS LLQGTILQYV KTLMEVMPKI C RLPRHEYG SPGILEFFHH QLKDIVEYAE LKTVCFQNLR EVGNAILFCL LIEQSLSLEE VCDLLHAAPF QNILPRVHVK EG ERLDAKM KRLESKYAPL HLVPLIERLG TPQQIAIARE GDLLTKERLC CGLSMFEVIL TRIRSFLDDP IWRGPLPSNG VMH VDECVE FHRLWSAMQF VYCIPVGTHE FTVEQCFGDG LHWAGCMIIV LLGQQRRFAV LDFCYHLLKV QKHDGKDEII KNVP LKKMV ERIRKFQILN DEIITILDKY LKSGDGEGTP VEHVRCFQPP IHQSLASS UniProtKB: Cytoplasmic FMR1-interacting protein 1 |
-分子 #2: Nck-associated protein 1
| 分子 | 名称: Nck-associated protein 1 / タイプ: protein_or_peptide / ID: 2 詳細: This construct contains two additional uncleaved residues "GA" in the N terminus from the construct design and purification procedure. Densities for these residues are not observed in the map ...詳細: This construct contains two additional uncleaved residues "GA" in the N terminus from the construct design and purification procedure. Densities for these residues are not observed in the map and were not included in the sample sequence to avoid numbering shifts. コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 128.940727 KDa |
| 組換発現 | 生物種:  Trichoplusia ni (イラクサキンウワバ) Trichoplusia ni (イラクサキンウワバ) |
| 配列 | 文字列: MSRSVLQPSQ QKLAEKLTIL NDRGVGMLTR LYNIKKACGD PKAKPSYLID KNLESAVKFI VRKFPAVETR NNNQQLAQLQ KEKSEILKN LALYYFTFVD VMEFKDHVCE LLNTIDVCQV FFDITVNFDL TKNYLDLIIT YTTLMILLSR IEERKAIIGL Y NYAHEMTH ...文字列: MSRSVLQPSQ QKLAEKLTIL NDRGVGMLTR LYNIKKACGD PKAKPSYLID KNLESAVKFI VRKFPAVETR NNNQQLAQLQ KEKSEILKN LALYYFTFVD VMEFKDHVCE LLNTIDVCQV FFDITVNFDL TKNYLDLIIT YTTLMILLSR IEERKAIIGL Y NYAHEMTH GASDREYPRL GQMIVDYENP LKKMMEEFVP HSKSLSDALI SLQMVYPRRN LSADQWRNAQ LLSLISAPST ML NPAQSDT MPCEYLSLDA MEKWIIFGFI LCHGILNTDA TALNLWKLAL QSSSCLSLFR DEVFHIHKAA EDLFVNIRGY NKR INDIRE CKEAAVSHAG SMHRERRKFL RSALKELATV LSDQPGLLGP KALFVFMALS FARDEIIWLL RHADNMPKKS ADDF IDKHI AELIFYMEEL RAHVRKYGPV MQRYYVQYLS GFDAVVLNEL VQNLSVCPED ESIIMSSFVN TMTSLSVKQV EDGEV FDFR GMRLDWFRLQ AYTSVSKASL GLADHRELGK MMNTIIFHTK MVDSLVEMLV ETSDLSIFCF YSRAFEKMFQ QCLELP SQS RYSIAFPLLC THFMSCTHEL CPEERHHIGD RSLSLCNMFL DEMAKQARNL ITDICTEQCT LSDQLLPKHC AKTISQA VN KKSKKQTGKK GEPEREKPGV ESMRKNRLVV TNLDKLHTAL SELCFSINYV PNMVVWEHTF TPREYLTSHL EIRFTKSI V GMTMYNQATQ EIAKPSELLT SVRAYMTVLQ SIENYVQIDI TRVFNNVLLQ QTQHLDSHGE PTITSLYTNW YLETLLRQV SNGHIAYFPA MKAFVNLPTE NELTFNAEEY SDISEMRSLS ELLGPYGMKF LSESLMWHIS SQVAELKKLV VENVDVLTQM RTSFDKPDQ MAALFKRLSS VDSVLKRMTI IGVILSFRSL AQEALRDVLS YHIPFLVSSI EDFKDHIPRE TDMKVAMNVY E LSSAAGLP CEIDPALVVA LSSQKSENIS PEEEYKIACL LMVFVAVSLP TLASNVMSQY SPAIEGHCNN IHCLAKAINQ IA AALFTIH KGSIEDRLKE FLALASSSLL KIGQETDKTT TRNRESVYLL LDMIVQESPF LTMDLLESCF PYVLLRNAYH AVY KQSVTS SA UniProtKB: Nck-associated protein 1 |
-分子 #3: Wiskott-Aldrich syndrome protein family member 1
| 分子 | 名称: Wiskott-Aldrich syndrome protein family member 1 / タイプ: protein_or_peptide / ID: 3 詳細: Residues 231-248 are inserted as a flexible linker sequence. This construct contains two additional uncleaved residues "GA" in the N terminus from the construct design and purification ...詳細: Residues 231-248 are inserted as a flexible linker sequence. This construct contains two additional uncleaved residues "GA" in the N terminus from the construct design and purification procedure. Densities for these residues are not observed in the map and were not included in the sample sequence to avoid numbering shifts. コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 37.009406 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MPLVKRNIDP RHLCHTALPR GIKNELECVT NISLANIIRQ LSSLSKYAED IFGELFNEAH SFSFRVNSLQ ERVDRLSVSV TQLDPKEEE LSLQDITMRK AFRSSTIQDQ QLFDRKTLPI PLQETYDVCE QPPPLNILTP YRDDGKEGLK FYTNPSYFFD L WKEKMLQD ...文字列: MPLVKRNIDP RHLCHTALPR GIKNELECVT NISLANIIRQ LSSLSKYAED IFGELFNEAH SFSFRVNSLQ ERVDRLSVSV TQLDPKEEE LSLQDITMRK AFRSSTIQDQ QLFDRKTLPI PLQETYDVCE QPPPLNILTP YRDDGKEGLK FYTNPSYFFD L WKEKMLQD TEDKRKEKRK QKQKNLDRPH EPEKVPRAPH DRRREWQKLA QGPELAEDDA NLLHKHIEVA NGGGSGGSGG SG GSGGSGG SKRHPSTLPV ISDARSVLLE AIRKGIQLRK VEEQREQEAK HERIENDVAT ILSRRIAVEY SDSEDDSEFD EVD WLE UniProtKB: Actin-binding protein WASF1, Actin-binding protein WASF1 |
-分子 #4: Protein BRICK1
| 分子 | 名称: Protein BRICK1 / タイプ: protein_or_peptide / ID: 4 詳細: This construct contains uncleaved residues "GHMGAA" in the N terminus from the construct design and purification procedure. Densities for the residues are not observed in the map and were not ...詳細: This construct contains uncleaved residues "GHMGAA" in the N terminus from the construct design and purification procedure. Densities for the residues are not observed in the map and were not included in the sample sequence to avoid numbering shifts. コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 8.756915 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MAGQEDPVQR EIHQDWANRE YIEIITSSIK KIADFLNSFD MSCRSRLATL NEKLTALERR IEYIEARVTK GETLT UniProtKB: Protein BRICK1 |
-分子 #5: Abl interactor 2
| 分子 | 名称: Abl interactor 2 / タイプ: protein_or_peptide / ID: 5 詳細: The sequence only contains residues 1-158. Also, there are two additional uncleaved residues "GH" in the N terminus from the construct design and purification procedure. Densities for these ...詳細: The sequence only contains residues 1-158. Also, there are two additional uncleaved residues "GH" in the N terminus from the construct design and purification procedure. Densities for these residues are not observed in the map and were not included in the sample sequence to avoid numbering shifts. コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 18.041482 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MAELQMLLEE EIPGGRRALF DSYTNLERVA DYCENNYIQS ADKQRALEET KAYTTQSLAS VAYLINTLAN NVLQMLDIQA SQLRRMESS INHISQTVDI HKEKVARREI GILTTNKNTS RTHKIIAPAN LERPVRYIRK PIDYTILDDI GHGVKVSTQ UniProtKB: Abl interactor 2 |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.41 mg/mL |
|---|---|
| 緩衝液 | pH: 7 |
| グリッド | モデル: UltrAuFoil R1.2/1.3 / 材質: GOLD / メッシュ: 300 / 支持フィルム - 材質: GOLD / 支持フィルム - トポロジー: HOLEY ARRAY / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 120 sec. / 前処理 - 雰囲気: AIR / 前処理 - 気圧: 0.019 kPa / 詳細: 30 mA |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 95 % / チャンバー内温度: 277.15 K / 装置: HOMEMADE PLUNGER |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TALOS ARCTICA |
|---|---|
| 詳細 | Data were collected by shifting the stage to the target exposure positions. |
| 撮影 | フィルム・検出器のモデル: FEI FALCON III (4k x 4k) 検出モード: COUNTING / デジタル化 - サイズ - 横: 4096 pixel / デジタル化 - サイズ - 縦: 4096 pixel / 撮影したグリッド数: 1 / 実像数: 2913 / 平均露光時間: 40.0 sec. / 平均電子線量: 44.06 e/Å2 詳細: Each micrograph was acquired as dose-fractionated movies consisting of 62 frames per movie. |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 50.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 1.2 µm / 最小 デフォーカス(公称値): 0.6 µm / 倍率(公称値): 120000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Talos Arctica / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)