+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of DHQS/EPSPS dimer from Candida albicans Aro1 | |||||||||
 Map data Map data | Sharpened map from focused refinement of DHQS/EPSPS region | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | BIOSYNTHETIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information3-dehydroquinate synthase / 3-dehydroquinate synthase activity / shikimate kinase / shikimate kinase activity / shikimate dehydrogenase (NADP+) / shikimate 3-dehydrogenase (NADP+) activity / 3-phosphoshikimate 1-carboxyvinyltransferase / 3-phosphoshikimate 1-carboxyvinyltransferase activity / 3-dehydroquinate dehydratase / 3-dehydroquinate dehydratase activity ...3-dehydroquinate synthase / 3-dehydroquinate synthase activity / shikimate kinase / shikimate kinase activity / shikimate dehydrogenase (NADP+) / shikimate 3-dehydrogenase (NADP+) activity / 3-phosphoshikimate 1-carboxyvinyltransferase / 3-phosphoshikimate 1-carboxyvinyltransferase activity / 3-dehydroquinate dehydratase / 3-dehydroquinate dehydratase activity / chorismate biosynthetic process / aromatic amino acid family biosynthetic process / amino acid biosynthetic process / ATP binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Candida albicans (yeast) Candida albicans (yeast) | |||||||||
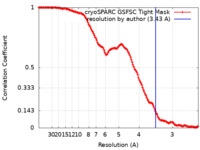
| Method | single particle reconstruction / cryo EM / Resolution: 3.43 Å | |||||||||
 Authors Authors | Quade B / Borek D / Otwinowski Z | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Life Sci Alliance / Year: 2022 Journal: Life Sci Alliance / Year: 2022Title: Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in . Authors: Peter J Stogios / Sean D Liston / Cameron Semper / Bradley Quade / Karolina Michalska / Elena Evdokimova / Shane Ram / Zbyszek Otwinowski / Dominika Borek / Leah E Cowen / Alexei Savchenko /   Abstract: In the human fungal pathogen , encodes an essential multi-enzyme that catalyses consecutive steps in the shikimate pathway for biosynthesis of chorismate, a precursor to folate and the aromatic ...In the human fungal pathogen , encodes an essential multi-enzyme that catalyses consecutive steps in the shikimate pathway for biosynthesis of chorismate, a precursor to folate and the aromatic amino acids. We obtained the first molecular image of Aro1 that reveals the architecture of all five enzymatic domains and their arrangement in the context of the full-length protein. Aro1 forms a flexible dimer allowing relative autonomy of enzymatic function of the individual domains. Our activity and in cellulo data suggest that only four of Aro1's enzymatic domains are functional and essential for viability of , whereas the 3-dehydroquinate dehydratase (DHQase) domain is inactive because of active site substitutions. We further demonstrate that in , the type II DHQase Dqd1 can compensate for the inactive DHQase domain of Aro1, suggesting an unrecognized essential role for this enzyme in shikimate biosynthesis. In contrast, in and , which do not encode a Dqd1 homolog, Aro1 DHQase domains are enzymatically active, highlighting diversity across species. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26358.map.gz emd_26358.map.gz | 230.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26358-v30.xml emd-26358-v30.xml emd-26358.xml emd-26358.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26358_fsc.xml emd_26358_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_26358.png emd_26358.png | 89.3 KB | ||
| Filedesc metadata |  emd-26358.cif.gz emd-26358.cif.gz | 6.9 KB | ||
| Others |  emd_26358_additional_1.map.gz emd_26358_additional_1.map.gz emd_26358_additional_2.map.gz emd_26358_additional_2.map.gz emd_26358_half_map_1.map.gz emd_26358_half_map_1.map.gz emd_26358_half_map_2.map.gz emd_26358_half_map_2.map.gz | 123.1 MB 213.6 MB 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26358 http://ftp.pdbj.org/pub/emdb/structures/EMD-26358 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26358 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26358 | HTTPS FTP |
-Validation report
| Summary document |  emd_26358_validation.pdf.gz emd_26358_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26358_full_validation.pdf.gz emd_26358_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_26358_validation.xml.gz emd_26358_validation.xml.gz | 21 KB | Display | |
| Data in CIF |  emd_26358_validation.cif.gz emd_26358_validation.cif.gz | 26.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26358 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26358 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26358 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26358 | HTTPS FTP |
-Related structure data
| Related structure data |  7u5tMC  6c5cC  7tbuC  7tbvC  7u5sC  7u5uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26358.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26358.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map from focused refinement of DHQS/EPSPS region | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2495 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Raw map from focused refinement of DHQS/EPSPS region
| File | emd_26358_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw map from focused refinement of DHQS/EPSPS region | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Postprocessed map of DHQS and EPSPS region from...
| File | emd_26358_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map of DHQS and EPSPS region from broken particles at 2.94A. Used for model building. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B from focused refinement of DHQS/EPSPS region
| File | emd_26358_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B from focused refinement of DHQS/EPSPS region | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A from focused refinement of DHQS/EPSPS region
| File | emd_26358_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A from focused refinement of DHQS/EPSPS region | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Aro1 from Candida albicans
| Entire | Name: Aro1 from Candida albicans |
|---|---|
| Components |
|
-Supramolecule #1: Aro1 from Candida albicans
| Supramolecule | Name: Aro1 from Candida albicans / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Candida albicans (yeast) / Strain: CaLC6830 Candida albicans (yeast) / Strain: CaLC6830 |
| Molecular weight | Theoretical: 169.4 KDa |
-Macromolecule #1: Pentafunctional AROM polypeptide
| Macromolecule | Name: Pentafunctional AROM polypeptide / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: 3-dehydroquinate synthase |
|---|---|
| Source (natural) | Organism:  Candida albicans (yeast) / Strain: SC5314 / ATCC MYA-2876 Candida albicans (yeast) / Strain: SC5314 / ATCC MYA-2876 |
| Molecular weight | Theoretical: 169.59625 KDa |
| Sequence | String: MSIEKVPILG KETIHVGYGI ADHIVREVIA NLASSTYVIV TDTNMARTPQ YSKLTDDFKT NLSEKRPESR LLTYCVSPGE NNKNRATKA AVEDFLLQQG CTRDTVILAV GGGVIGDMIG FVAATFMRGV RVVQVPTTLL AMVDSSVGGK TAIDTPLGKN F IGAFHQPE ...String: MSIEKVPILG KETIHVGYGI ADHIVREVIA NLASSTYVIV TDTNMARTPQ YSKLTDDFKT NLSEKRPESR LLTYCVSPGE NNKNRATKA AVEDFLLQQG CTRDTVILAV GGGVIGDMIG FVAATFMRGV RVVQVPTTLL AMVDSSVGGK TAIDTPLGKN F IGAFHQPE YVFCDVSFLE TLPARQFING MAEVVKTAAI WNEEEFTRLE NFSKKFLSVV TSKKPDLQSI KAELVKTVLE SV RVKAGVV SSDEKEAGLR NLLNFGHTIG HAIEAVLTPE ALHGECVSIG MIKEAELSRY LGILPPVAVA RLSKCLVAYG LPV SIDDKE FLKKVGPKRH YVEIDILLKK MAIDKKNDGS KIRCVLLEKI GKCYQLKAHQ VSKQDLSFVL TDEVLVHPFT NPPK ENIIV PPGSKSISNR ALILAALGNG TVRVKNLLHS DDTKHMLDAV ASLKGAEIST EDNGETIVVK GNGGNLVTSG EELYL GNAG TASRFLTTVA SLVGKSQASD DVILTGNARM QERPIGPLVD ALGSNGSEIE YLNKQGSLPL KISAGNGLKG GRIELA ATI SSQYVSSILM CAPYAKEPVT LALVGGKPIS QLYIDMTCAM MKSFGIEVTK STTEEYTYHI PKGTYKNPSE YVIESDA SS ATYPLAFAAM TGTSCTIPNI GSSSLQGDAK FAVDVLKPMG CKVEQTTTST TVTGPPRGHL KPLPHVDMEP MTDAFLTA S VVAAVAKGGS STSITGIANQ RVKECNRIEA MVTELAKFGV PANELPDGIE IHGIDIEDLK TPEISKRGVS SYDDHRVAM SFSLLAGLCK EPVLILERST TGKTWPGWWD ILHSKFKIEL DGYEPPFNTD KHVDKSSDKS IIVIGMRGTG KSTLSEWLAS FLGFKMLDM DKYLEEKLGT GIKSLIKAKG WEYFRQEEAI VAKECFTKFS KGYVLSTGGG IVEGEDARQQ LKSYADNGGI V LHLHRDLD ETVTFLAADT TRPAYSSEVQ EVWLRREKWY HECSNYHFYS SHCSTEDEFN HLRRSFVNYI KLITGAERPV VP AGRSAAV VLTSPDLNEV VGDLESITIG ADAVELRVDL FKDTSAEFVA AQIAVIRKHA DLPIIYTVRT VSQGGKFPDE NVD ELKSLL LLGIRLGVAY VDLQLTAPNE LIEEISSKKG FTRVIGTYQD INGELKWNNV EWKNKYNQGV SMNADIVRLV GKAN SIQDN LDLENFKKQN TLKPLIAFNL GSQGKLSQVL NGTFTPISHK LLPNDEEFLT IGELNQTYFD IGGFTAKKFW VIGSP IEHS RSPNLHNAGY KALNLPYQFG RFEATDVDVV YDNLINKPDF GGLAITMPLK LDIMKFATKL SDAAETIGAV NTLIPI EGG YFGDNTDWVG ISNSFIRAGV PPKSSSNGLV VGAGGTSRAA IYALHQMGCA KIYLVNRTAA KLEELVKSFP KDYNLEI VE TEQQADKASK VSLAVSCIPA DKPLDGEVLK KIERILSNGS EQSAGFKPTL LEASYKPRVT PIMKLTEEQY KWKVIPGV E MLVNQGDRQF KLHTGFTAPY EIIHRAVVEE UniProtKB: Pentafunctional AROM polypeptide |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 80 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 83.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||||
| Output model |  PDB-7u5t: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)