[English] 日本語
 Yorodumi
Yorodumi- EMDB-26333: Cryo-EM structure of centromeric chromatin array shaped by the ki... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of centromeric chromatin array shaped by the kinetochore protein CENP-N | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
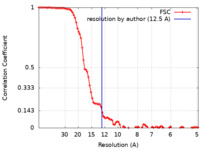
| Method | single particle reconstruction / cryo EM / Resolution: 12.5 Å | |||||||||
 Authors Authors | Zhou K / Luger K | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: CENP-N promotes the compaction of centromeric chromatin. Authors: Keda Zhou / Magdalena Gebala / Dustin Woods / Kousik Sundararajan / Garrett Edwards / Dan Krzizike / Jeff Wereszczynski / Aaron F Straight / Karolin Luger /  Abstract: The histone variant CENP-A is the epigenetic determinant for the centromere, where it is interspersed with canonical H3 to form a specialized chromatin structure that nucleates the kinetochore. How ...The histone variant CENP-A is the epigenetic determinant for the centromere, where it is interspersed with canonical H3 to form a specialized chromatin structure that nucleates the kinetochore. How nucleosomes at the centromere arrange into higher order structures is unknown. Here we demonstrate that the human CENP-A-interacting protein CENP-N promotes the stacking of CENP-A-containing mononucleosomes and nucleosomal arrays through a previously undefined interaction between the α6 helix of CENP-N with the DNA of a neighboring nucleosome. We describe the cryo-EM structures and biophysical characterization of such CENP-N-mediated nucleosome stacks and nucleosomal arrays and demonstrate that this interaction is responsible for the formation of densely packed chromatin at the centromere in the cell. Our results provide first evidence that CENP-A, together with CENP-N, promotes specific chromatin higher order structure at the centromere. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26333.map.gz emd_26333.map.gz | 59.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26333-v30.xml emd-26333-v30.xml emd-26333.xml emd-26333.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26333_fsc.xml emd_26333_fsc.xml | 9.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_26333.png emd_26333.png | 30.2 KB | ||
| Masks |  emd_26333_msk_1.map emd_26333_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_26333_half_map_1.map.gz emd_26333_half_map_1.map.gz emd_26333_half_map_2.map.gz emd_26333_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26333 http://ftp.pdbj.org/pub/emdb/structures/EMD-26333 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26333 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26333 | HTTPS FTP |
-Validation report
| Summary document |  emd_26333_validation.pdf.gz emd_26333_validation.pdf.gz | 919.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26333_full_validation.pdf.gz emd_26333_full_validation.pdf.gz | 918.6 KB | Display | |
| Data in XML |  emd_26333_validation.xml.gz emd_26333_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_26333_validation.cif.gz emd_26333_validation.cif.gz | 21.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26333 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26333 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26333 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26333 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26333.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26333.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.48 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26333_msk_1.map emd_26333_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26333_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_26333_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Centromeric chromatin array in complex with kinetochore protein CENP-N
| Entire | Name: Centromeric chromatin array in complex with kinetochore protein CENP-N |
|---|---|
| Components |
|
-Supramolecule #1: Centromeric chromatin array in complex with kinetochore protein CENP-N
| Supramolecule | Name: Centromeric chromatin array in complex with kinetochore protein CENP-N type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 100.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)