+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
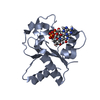
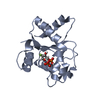
| タイトル | Cryo-EM structure of human SARM1 TIR domain in complex with 1AD | |||||||||
 マップデータ マップデータ | Main map of hSARM1-TIR : 1AD complex | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | NADase / Axon degeneration / Inhibitor / Complex / CYTOSOLIC PROTEIN | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
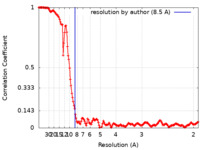
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 8.5 Å | |||||||||
 データ登録者 データ登録者 | Saikot FK / Kobe B / Ve T / Nanson JD / Gu W / Luo Z / Brillault L / Landsberg MJ | |||||||||
| 資金援助 |  オーストラリア, 1件 オーストラリア, 1件
| |||||||||
 引用 引用 |  ジャーナル: Mol Cell / 年: 2022 ジャーナル: Mol Cell / 年: 2022タイトル: Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules. 著者: Yun Shi / Philip S Kerry / Jeffrey D Nanson / Todd Bosanac / Yo Sasaki / Raul Krauss / Forhad K Saikot / Sarah E Adams / Tamim Mosaiab / Veronika Masic / Xianrong Mao / Faith Rose / Eduardo ...著者: Yun Shi / Philip S Kerry / Jeffrey D Nanson / Todd Bosanac / Yo Sasaki / Raul Krauss / Forhad K Saikot / Sarah E Adams / Tamim Mosaiab / Veronika Masic / Xianrong Mao / Faith Rose / Eduardo Vasquez / Marieke Furrer / Katie Cunnea / Andrew Brearley / Weixi Gu / Zhenyao Luo / Lou Brillault / Michael J Landsberg / Aaron DiAntonio / Bostjan Kobe / Jeffrey Milbrandt / Robert O Hughes / Thomas Ve /     要旨: The NADase SARM1 (sterile alpha and TIR motif containing 1) is a key executioner of axon degeneration and a therapeutic target for several neurodegenerative conditions. We show that a potent SARM1 ...The NADase SARM1 (sterile alpha and TIR motif containing 1) is a key executioner of axon degeneration and a therapeutic target for several neurodegenerative conditions. We show that a potent SARM1 inhibitor undergoes base exchange with the nicotinamide moiety of nicotinamide adenine dinucleotide (NAD) to produce the bona fide inhibitor 1AD. We report structures of SARM1 in complex with 1AD, NAD mimetics and the allosteric activator nicotinamide mononucleotide (NMN). NMN binding triggers reorientation of the armadillo repeat (ARM) domains, which disrupts ARM:TIR interactions and leads to formation of a two-stranded TIR domain assembly. The active site spans two molecules in these assemblies, explaining the requirement of TIR domain self-association for NADase activity and axon degeneration. Our results reveal the mechanisms of SARM1 activation and substrate binding, providing rational avenues for the design of new therapeutics targeting SARM1. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_26191.map.gz emd_26191.map.gz | 170 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-26191-v30.xml emd-26191-v30.xml emd-26191.xml emd-26191.xml | 19.4 KB 19.4 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_26191_fsc.xml emd_26191_fsc.xml | 15.6 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_26191.png emd_26191.png | 31.9 KB | ||
| マスクデータ |  emd_26191_msk_1.map emd_26191_msk_1.map | 347.6 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-26191.cif.gz emd-26191.cif.gz | 5.3 KB | ||
| その他 |  emd_26191_additional_1.map.gz emd_26191_additional_1.map.gz emd_26191_half_map_1.map.gz emd_26191_half_map_1.map.gz emd_26191_half_map_2.map.gz emd_26191_half_map_2.map.gz | 173 MB 323 MB 323 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26191 http://ftp.pdbj.org/pub/emdb/structures/EMD-26191 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26191 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26191 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_26191_validation.pdf.gz emd_26191_validation.pdf.gz | 1.2 MB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_26191_full_validation.pdf.gz emd_26191_full_validation.pdf.gz | 1.2 MB | 表示 | |
| XML形式データ |  emd_26191_validation.xml.gz emd_26191_validation.xml.gz | 24.1 KB | 表示 | |
| CIF形式データ |  emd_26191_validation.cif.gz emd_26191_validation.cif.gz | 31.2 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26191 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26191 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26191 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26191 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_26191.map.gz / 形式: CCP4 / 大きさ: 347.6 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_26191.map.gz / 形式: CCP4 / 大きさ: 347.6 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Main map of hSARM1-TIR : 1AD complex | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.96 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_26191_msk_1.map emd_26191_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-追加マップ: Sharpened map of hSARM1-TIR : 1AD complex
| ファイル | emd_26191_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Sharpened map of hSARM1-TIR : 1AD complex | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Half A volume map of hSARM1-TIR : 1AD complex
| ファイル | emd_26191_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half A volume map of hSARM1-TIR : 1AD complex | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Half B volume map of hSARM1-TIR : 1AD complex
| ファイル | emd_26191_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half B volume map of hSARM1-TIR : 1AD complex | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Active state assembly of hSARM1-TIR domain with 1AD.
| 全体 | 名称: Active state assembly of hSARM1-TIR domain with 1AD. |
|---|---|
| 要素 |
|
-超分子 #1: Active state assembly of hSARM1-TIR domain with 1AD.
| 超分子 | 名称: Active state assembly of hSARM1-TIR domain with 1AD. タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: hSARM1-TIR domain : 1AD complex
| 分子 | 名称: hSARM1-TIR domain : 1AD complex / タイプ: other / ID: 1 詳細: 1AD = Base exchanged product of NAD+ and 5-iodoisoquinoline 分類: other |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: SNATPDVFIS YRRNSGSQLA SLLKVHLQLH GFSVFIDVEK LEAGKFEDKL IQSVMGARNF VLVLSPGALD KCMQDHDCKD WVHKEIVTAL SCGKNIVPII DGFEWPEPQV LPEDMQAVLT FNGIKWSHEY QEATIEKIIR FLQ |
| 組換発現 | 生物種:  |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 3 mg/mL |
|---|---|
| 緩衝液 | pH: 7.5 |
| グリッド | モデル: Quantifoil R2/2 / 材質: GOLD / メッシュ: 400 / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 60 sec. / 前処理 - 雰囲気: OTHER |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 95 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK IV |
| 詳細 | This sample was oligomer. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | JEOL CRYO ARM 300 |
|---|---|
| 特殊光学系 | エネルギーフィルター - 名称: In-column Omega Filter エネルギーフィルター - スリット幅: 20 eV |
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 撮影したグリッド数: 1 / 実像数: 247 / 平均露光時間: 5.0 sec. / 平均電子線量: 65.1 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: OTHER / 撮影モード: OTHER / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 0.5 µm / 倍率(公称値): 50000 |
| 試料ステージ | 試料ホルダーモデル: JEOL CRYOSPECPORTER / ホルダー冷却材: NITROGEN |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | プロトコル: RIGID BODY FIT |
|---|
 ムービー
ムービー コントローラー
コントローラー











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)