[English] 日本語
 Yorodumi
Yorodumi- EMDB-25995: TMEM106B singlet filament extracted from MSTD neurodegenerative h... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TMEM106B singlet filament extracted from MSTD neurodegenerative human brain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TMEM106B / TMEM filament / MSTD / NEUROPEPTIDE | |||||||||
| Function / homology |  Function and homology information Function and homology informationlysosomal protein catabolic process / regulation of lysosome organization / lysosomal lumen acidification / lysosome localization / lysosomal transport / positive regulation of dendrite development / dendrite morphogenesis / lysosome organization / neuron cellular homeostasis / late endosome membrane ...lysosomal protein catabolic process / regulation of lysosome organization / lysosomal lumen acidification / lysosome localization / lysosomal transport / positive regulation of dendrite development / dendrite morphogenesis / lysosome organization / neuron cellular homeostasis / late endosome membrane / ATPase binding / lysosome / endosome / lysosomal membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.25 Å | |||||||||
 Authors Authors | Hoq MR / Bharath SR | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Acta Neuropathol / Year: 2023 Journal: Acta Neuropathol / Year: 2023Title: Cross-β helical filaments of Tau and TMEM106B in gray and white matter of multiple system tauopathy with presenile dementia. Authors: Md Rejaul Hoq / Sakshibeedu R Bharath / Grace I Hallinan / Anllely Fernandez / Frank S Vago / Kadir A Ozcan / Daoyi Li / Holly J Garringer / Ruben Vidal / Bernardino Ghetti / Wen Jiang /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25995.map.gz emd_25995.map.gz | 99.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25995-v30.xml emd-25995-v30.xml emd-25995.xml emd-25995.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25995.png emd_25995.png | 140.7 KB | ||
| Masks |  emd_25995_msk_1.map emd_25995_msk_1.map emd_25995_msk_2.map emd_25995_msk_2.map | 67 MB 67 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25995.cif.gz emd-25995.cif.gz | 5.8 KB | ||
| Others |  emd_25995_half_map_1.map.gz emd_25995_half_map_1.map.gz emd_25995_half_map_2.map.gz emd_25995_half_map_2.map.gz | 47 MB 47 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25995 http://ftp.pdbj.org/pub/emdb/structures/EMD-25995 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25995 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25995 | HTTPS FTP |
-Validation report
| Summary document |  emd_25995_validation.pdf.gz emd_25995_validation.pdf.gz | 963.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25995_full_validation.pdf.gz emd_25995_full_validation.pdf.gz | 963.5 KB | Display | |
| Data in XML |  emd_25995_validation.xml.gz emd_25995_validation.xml.gz | 14.4 KB | Display | |
| Data in CIF |  emd_25995_validation.cif.gz emd_25995_validation.cif.gz | 16.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25995 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25995 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25995 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25995 | HTTPS FTP |
-Related structure data
| Related structure data |  7tmcMC  8f9kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25995.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25995.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.054 Å | ||||||||||||||||||||||||||||||||||||
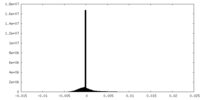
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25995_msk_1.map emd_25995_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
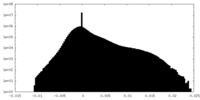
| Density Histograms |
-Mask #2
| File |  emd_25995_msk_2.map emd_25995_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25995_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_25995_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TMEM106B
| Entire | Name: TMEM106B |
|---|---|
| Components |
|
-Supramolecule #1: TMEM106B
| Supramolecule | Name: TMEM106B / type: tissue / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transmembrane protein 106B
| Macromolecule | Name: Transmembrane protein 106B / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.156318 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGKSLSHLPL HSSKEDAYDG VTSENMRNGL VNSEVHNEDG RNGDVSQFPY VEFTGRDSVT CPTCQGTGRI PRGQENQLVA LIPYSDQRL RPRRTKLYVM ASVFVCLLLS GLAVFFLFPR SIDVKYIGVK SAYVSYDVQK RTIYLNITNT LNITNNNYYS V EVENITAQ ...String: MGKSLSHLPL HSSKEDAYDG VTSENMRNGL VNSEVHNEDG RNGDVSQFPY VEFTGRDSVT CPTCQGTGRI PRGQENQLVA LIPYSDQRL RPRRTKLYVM ASVFVCLLLS GLAVFFLFPR SIDVKYIGVK SAYVSYDVQK RTIYLNITNT LNITNNNYYS V EVENITAQ VQFSKTVIGK ARLNNITIIG PLDMKQIDYT VPTVIAEEMS YMYDFCTLIS IKVHNIVLMM QVTVTTTYFG HS EQISQER YQYVDCGRNT TYQLGQSEYL NVLQPQQ UniProtKB: Transmembrane protein 106B |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.2 |
| Grid | Model: C-flat-1.2/1.3 / Material: GRAPHENE OXIDE / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 1.103 sec. / Average electron dose: 50.46 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number classes used: 1 Applied symmetry - Helical parameters - Δz: 4.79 Å Applied symmetry - Helical parameters - Δ&Phi: -0.43 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 3.25 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 16484 |
|---|---|
| Startup model | Type of model: NONE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller



 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)