[English] 日本語
 Yorodumi
Yorodumi- EMDB-23995: CryoEM Structure of Full-Length mGlu2 Bound to Ago-PAM ADX55164 a... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23995 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM Structure of Full-Length mGlu2 Bound to Ago-PAM ADX55164 and Glutamate | |||||||||
 Map data Map data | CryoEM Map of Full-Length mGlu2 Bound to Ago-PAM ADX55164 and Glutamate | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Metabotropic Glutamate Receptor 2 (mGlu2) (mGluR2) / Family C G protein-coupled receptor (GPCR) / Heterotrimeric G protein / CryoEM structure / MEMBRANE PROTEIN / MEMBRANE PROTEIN-AGONIST complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of response to drug / group II metabotropic glutamate receptor activity / intracellular glutamate homeostasis / behavioral response to nicotine / negative regulation of adenylate cyclase activity / G protein-coupled glutamate receptor signaling pathway / astrocyte projection / Class C/3 (Metabotropic glutamate/pheromone receptors) / glutamate secretion / glutamate receptor activity ...regulation of response to drug / group II metabotropic glutamate receptor activity / intracellular glutamate homeostasis / behavioral response to nicotine / negative regulation of adenylate cyclase activity / G protein-coupled glutamate receptor signaling pathway / astrocyte projection / Class C/3 (Metabotropic glutamate/pheromone receptors) / glutamate secretion / glutamate receptor activity / regulation of glutamate secretion / long-term synaptic depression / regulation of dopamine secretion / calcium channel regulator activity / regulation of synaptic transmission, glutamatergic / presynaptic modulation of chemical synaptic transmission / response to cocaine / G protein-coupled receptor activity / presynaptic membrane / gene expression / G alpha (i) signalling events / scaffold protein binding / chemical synaptic transmission / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / axon / glutamatergic synapse / dendrite / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Seven AB / Barros-Alvarez X | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: G-protein activation by a metabotropic glutamate receptor. Authors: Alpay B Seven / Ximena Barros-Álvarez / Marine de Lapeyrière / Makaía M Papasergi-Scott / Michael J Robertson / Chensong Zhang / Robert M Nwokonko / Yang Gao / Justin G Meyerowitz / Jean- ...Authors: Alpay B Seven / Ximena Barros-Álvarez / Marine de Lapeyrière / Makaía M Papasergi-Scott / Michael J Robertson / Chensong Zhang / Robert M Nwokonko / Yang Gao / Justin G Meyerowitz / Jean-Philippe Rocher / Dominik Schelshorn / Brian K Kobilka / Jesper M Mathiesen / Georgios Skiniotis /    Abstract: Family C G-protein-coupled receptors (GPCRs) operate as obligate dimers with extracellular domains that recognize small ligands, leading to G-protein activation on the transmembrane (TM) domains of ...Family C G-protein-coupled receptors (GPCRs) operate as obligate dimers with extracellular domains that recognize small ligands, leading to G-protein activation on the transmembrane (TM) domains of these receptors by an unknown mechanism. Here we show structures of homodimers of the family C metabotropic glutamate receptor 2 (mGlu2) in distinct functional states and in complex with heterotrimeric G. Upon activation of the extracellular domain, the two transmembrane domains undergo extensive rearrangement in relative orientation to establish an asymmetric TM6-TM6 interface that promotes conformational changes in the cytoplasmic domain of one protomer. Nucleotide-bound G can be observed pre-coupled to inactive mGlu2, but its transition to the nucleotide-free form seems to depend on establishing the active-state TM6-TM6 interface. In contrast to family A and B GPCRs, G-protein coupling does not involve the cytoplasmic opening of TM6 but is facilitated through the coordination of intracellular loops 2 and 3, as well as a critical contribution from the C terminus of the receptor. The findings highlight the synergy of global and local conformational transitions to facilitate a new mode of G-protein activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23995.map.gz emd_23995.map.gz | 296.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23995-v30.xml emd-23995-v30.xml emd-23995.xml emd-23995.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23995.png emd_23995.png | 98.2 KB | ||
| Filedesc metadata |  emd-23995.cif.gz emd-23995.cif.gz | 6.2 KB | ||
| Others |  emd_23995_additional_1.map.gz emd_23995_additional_1.map.gz | 320.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23995 http://ftp.pdbj.org/pub/emdb/structures/EMD-23995 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23995 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23995 | HTTPS FTP |
-Validation report
| Summary document |  emd_23995_validation.pdf.gz emd_23995_validation.pdf.gz | 305.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23995_full_validation.pdf.gz emd_23995_full_validation.pdf.gz | 304.8 KB | Display | |
| Data in XML |  emd_23995_validation.xml.gz emd_23995_validation.xml.gz | 7.5 KB | Display | |
| Data in CIF |  emd_23995_validation.cif.gz emd_23995_validation.cif.gz | 8.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23995 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23995 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23995 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23995 | HTTPS FTP |
-Related structure data
| Related structure data |  7mtrMC  7mtqC  7mtsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10978 (Title: CryoEM Structure of Full-Length mGlu2 Bound to Ago-PAM ADX55164 and Glutamate EMPIAR-10978 (Title: CryoEM Structure of Full-Length mGlu2 Bound to Ago-PAM ADX55164 and GlutamateData size: 3.5 TB Data #1: Unaligned multi-frame micrographs of Full-Length mGlu2 Bound to Ago-PAM ADX55164 and Glutamate [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23995.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23995.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM Map of Full-Length mGlu2 Bound to Ago-PAM ADX55164 and Glutamate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8677 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Local and focused refinement of mGlu2 CRD and 7TM domains
| File | emd_23995_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local and focused refinement of mGlu2 CRD and 7TM domains | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Full-length mGlu2 in ago-PAM and L-Glutamate bound-state
| Entire | Name: Full-length mGlu2 in ago-PAM and L-Glutamate bound-state |
|---|---|
| Components |
|
-Supramolecule #1: Full-length mGlu2 in ago-PAM and L-Glutamate bound-state
| Supramolecule | Name: Full-length mGlu2 in ago-PAM and L-Glutamate bound-state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Metabotropic glutamate receptor 2 bound to ago-PAM ADX55164 and L-Glutamate |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 191 KDa |
-Macromolecule #1: Metabotropic glutamate receptor 2
| Macromolecule | Name: Metabotropic glutamate receptor 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.932445 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AEGPAKKVLT LEGDLVLGGL FPVHQKGGPA EDCGPVNEHR GIQRLEAMLF ALDRINRDPH LLPGVRLGAH ILDSCSKDTH ALEQALDFV RASLSRGADG SRHICPDGSY ATHGDAPTAI TGVIGGSYSD VSIQVANLLR LFQIPQISYA STSAKLSDKS R YDYFARTV ...String: AEGPAKKVLT LEGDLVLGGL FPVHQKGGPA EDCGPVNEHR GIQRLEAMLF ALDRINRDPH LLPGVRLGAH ILDSCSKDTH ALEQALDFV RASLSRGADG SRHICPDGSY ATHGDAPTAI TGVIGGSYSD VSIQVANLLR LFQIPQISYA STSAKLSDKS R YDYFARTV PPDFFQAKAM AEILRFFNWT YVSTVASEGD YGETGIEAFE LEARARNICV ATSEKVGRAM SRAAFEGVVR AL LQKPSAR VAVLFTRSED ARELLAASQR LNASFTWVAS DGWGALESVV AGSEGAAEGA ITIELASYPI SDFASYFQSL DPW NNSRNP WFREFWEQRF RCSFRQRDCA AHSLRAVPFE QESKIMFVVN AVYAMAHALH NMHRALCPNT TRLCDAMRPV NGRR LYKDF VLNVKFDAPF RPADTHNEVR FDRFGDGIGR YNIFTYLRAG SGRYRYQKVG YWAEGLTLDT SLIPWASPSA GPLPA SRCS EPCLQNEVKS VQPGEVCCWL CIPCQPYEYR LDEFTCADCG LGYWPNASLT GCFELPQEYI RWGDAWAVGP VTIACL GAL ATLFVLGVFV RHNATPVVKA SGRELCYILL GGVFLCYCMT FIFIAKPSTA VCTLRRLGLG TAFSVCYSAL LTKTNRI AR IFGGAREGAQ RPRFISPASQ VAICLALISG QLLIVVAWLV VEAPGTGKET APERREVVTL RCNHRDASML GSLAYNVL L IALCTLYAFK TRKCPENFNE AKFIGFTMYT TCIIWLAFLP IFYVTSSDYR VQTTTMCVSV SLSGSVVLGC LFAPKLHII LFQPQKNVVS HRAPTSRFGS AAARASSSLG QGSGSQFVPT VCNGREVVDS TTSSL UniProtKB: Metabotropic glutamate receptor 2 |
-Macromolecule #2: GLUTAMIC ACID
| Macromolecule | Name: GLUTAMIC ACID / type: ligand / ID: 2 / Number of copies: 2 / Formula: GLU |
|---|---|
| Molecular weight | Theoretical: 147.129 Da |
| Chemical component information |  ChemComp-GLU: |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: 2-methoxy-6-propyl-N-(2-{4-[(1H-tetrazol-5-yl)methoxy]phenyl}ethy...
| Macromolecule | Name: 2-methoxy-6-propyl-N-(2-{4-[(1H-tetrazol-5-yl)methoxy]phenyl}ethyl)thieno[2,3-d]pyrimidin-4-amine type: ligand / ID: 4 / Number of copies: 1 / Formula: ZQY |
|---|---|
| Molecular weight | Theoretical: 425.507 Da |
| Chemical component information | 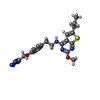 ChemComp-ZQY: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: GOLD / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.2) / Number images used: 219019 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)