[English] 日本語
 Yorodumi
Yorodumi- EMDB-23589: Cryo-EM structure of 2909 Fab in complex with 3BNC117 Fab and CAP... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23589 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of 2909 Fab in complex with 3BNC117 Fab and CAP256.wk34.c80 SOSIP.RnS2 N160K HIV-1 Env trimer | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CAP256 / V1V2 / V2-Apex / SOSIP / Vaccine / TYS / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / virus-mediated perturbation of host defense response / fusion of virus membrane with host endosome membrane / viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / virus-mediated perturbation of host defense response / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / apoptotic process / host cell plasma membrane / structural molecule activity / virion membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.91 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Extended antibody-framework-to-antigen distance observed exclusively with broad HIV-1-neutralizing antibodies recognizing glycan-dense surfaces. Authors: Myungjin Lee / Anita Changela / Jason Gorman / Reda Rawi / Tatsiana Bylund / Cara W Chao / Bob C Lin / Mark K Louder / Adam S Olia / Baoshan Zhang / Nicole A Doria-Rose / Susan Zolla-Pazner ...Authors: Myungjin Lee / Anita Changela / Jason Gorman / Reda Rawi / Tatsiana Bylund / Cara W Chao / Bob C Lin / Mark K Louder / Adam S Olia / Baoshan Zhang / Nicole A Doria-Rose / Susan Zolla-Pazner / Lawrence Shapiro / Gwo-Yu Chuang / Peter D Kwong /  Abstract: Antibody-Framework-to-Antigen Distance (AFAD) - the distance between the body of an antibody and a protein antigen - is an important parameter governing antibody recognition. Here, we quantify AFAD ...Antibody-Framework-to-Antigen Distance (AFAD) - the distance between the body of an antibody and a protein antigen - is an important parameter governing antibody recognition. Here, we quantify AFAD for ~2,000 non-redundant antibody-protein-antigen complexes in the Protein Data Bank. AFADs showed a gaussian distribution with mean of 16.3 Å and standard deviation (σ) of 2.4 Å. Notably, antibody-antigen complexes with extended AFADs (>3σ) were exclusively human immunodeficiency virus-type 1 (HIV-1)-neutralizing antibodies. High correlation (R = 0.8110) was observed between AFADs and glycan coverage, as assessed by molecular dynamics simulations of the HIV-1-envelope trimer. Especially long AFADs were observed for antibodies targeting the glycosylated trimer apex, and we tested the impact of introducing an apex-glycan hole (N160K); the cryo-EM structure of the glycan hole-targeting HIV-1-neutralizing antibody 2909 in complex with an N160K-envelope trimer revealed a substantially shorter AFAD. Overall, extended AFADs exclusively recognized densely glycosylated surfaces, with the introduction of a glycan hole enabling closer recognition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23589.map.gz emd_23589.map.gz | 228.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23589-v30.xml emd-23589-v30.xml emd-23589.xml emd-23589.xml | 26.2 KB 26.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23589.png emd_23589.png | 108.2 KB | ||
| Masks |  emd_23589_msk_1.map emd_23589_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23589.cif.gz emd-23589.cif.gz | 7.4 KB | ||
| Others |  emd_23589_additional_1.map.gz emd_23589_additional_1.map.gz emd_23589_additional_2.map.gz emd_23589_additional_2.map.gz emd_23589_half_map_1.map.gz emd_23589_half_map_1.map.gz emd_23589_half_map_2.map.gz emd_23589_half_map_2.map.gz | 14.6 MB 63.9 MB 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23589 http://ftp.pdbj.org/pub/emdb/structures/EMD-23589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23589 | HTTPS FTP |
-Validation report
| Summary document |  emd_23589_validation.pdf.gz emd_23589_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23589_full_validation.pdf.gz emd_23589_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_23589_validation.xml.gz emd_23589_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_23589_validation.cif.gz emd_23589_validation.cif.gz | 19.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23589 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23589 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23589 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23589 | HTTPS FTP |
-Related structure data
| Related structure data |  7ly9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23589.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23589.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0961 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
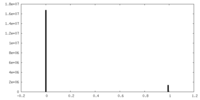
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23589_msk_1.map emd_23589_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
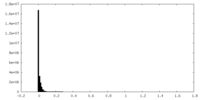
| Density Histograms |
-Additional map: Density modified map from phenix resolve
| File | emd_23589_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density modified map from phenix resolve | ||||||||||||
| Projections & Slices |
| ||||||||||||
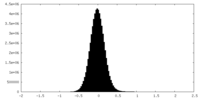
| Density Histograms |
-Additional map: Unsharpened Map
| File | emd_23589_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
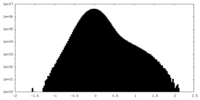
| Density Histograms |
-Half map: Half map B
| File | emd_23589_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
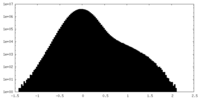
| Density Histograms |
-Half map: Half map A
| File | emd_23589_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 2909 Fab in complex with 3BNC117 Fab and CAP256.wk34.c80 SOSIP.Rn...
| Entire | Name: 2909 Fab in complex with 3BNC117 Fab and CAP256.wk34.c80 SOSIP.RnS2 N160K HIV-1 Env trimer |
|---|---|
| Components |
|
-Supramolecule #1: 2909 Fab in complex with 3BNC117 Fab and CAP256.wk34.c80 SOSIP.Rn...
| Supramolecule | Name: 2909 Fab in complex with 3BNC117 Fab and CAP256.wk34.c80 SOSIP.RnS2 N160K HIV-1 Env trimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: 2909 Light Chain
| Macromolecule | Name: 2909 Light Chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.582885 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SYVLTQPPSV SVAPGKTARI TCGGNNIANK NVHWYQQKPG QAPVLVIYYD DDRPSGIPDR FSGSNSGNTA TLTISRVEAG DEADYYCQV WDSNSDHVVF GGGTQLTVLG QPKAAPSVTL FPPSSEELQA NKATLVCLIS DFYPGAVTVA WKADSSPVKA G VETTTPSK ...String: SYVLTQPPSV SVAPGKTARI TCGGNNIANK NVHWYQQKPG QAPVLVIYYD DDRPSGIPDR FSGSNSGNTA TLTISRVEAG DEADYYCQV WDSNSDHVVF GGGTQLTVLG QPKAAPSVTL FPPSSEELQA NKATLVCLIS DFYPGAVTVA WKADSSPVKA G VETTTPSK QSNNKYAASS YLSLTPEQWK SHRSYSCQVT HEGSTVEKTV APT |
-Macromolecule #2: 2909 Heavy Chain
| Macromolecule | Name: 2909 Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.067795 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGN VVQPGGSLRL SCTASGFSFD DSTMHWVRQA PGKGLQWVSL ISWNGGRTYY ADSVKGRFTI SRDNSKNSLY LQMNSLKTE DTAFYFCAKD KGDSD(TYS)D(TYS)NL GYSYFYYMDG WGKGTTVTVS SASTKGPSVF PLAPSSKSTS GGT AALGCL ...String: EVQLVESGGN VVQPGGSLRL SCTASGFSFD DSTMHWVRQA PGKGLQWVSL ISWNGGRTYY ADSVKGRFTI SRDNSKNSLY LQMNSLKTE DTAFYFCAKD KGDSD(TYS)D(TYS)NL GYSYFYYMDG WGKGTTVTVS SASTKGPSVF PLAPSSKSTS GGT AALGCL VKDYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKRV EPK |
-Macromolecule #3: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.337512 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GLWVTVYYGV PVWREAKTTL FCASDAKSYE KEVHNVWATH ACVPTDPNPQ ELVLENVTEN FNMWKNDMVD QMHEDIISLW DQSLKPCVK LTPLCVTLNC SDAKVNATYK GTREEIKNCS FKATTELRDK KRREYALFYR LDIVPLSGEG NNNSEYRLIN C NTSVITQI ...String: GLWVTVYYGV PVWREAKTTL FCASDAKSYE KEVHNVWATH ACVPTDPNPQ ELVLENVTEN FNMWKNDMVD QMHEDIISLW DQSLKPCVK LTPLCVTLNC SDAKVNATYK GTREEIKNCS FKATTELRDK KRREYALFYR LDIVPLSGEG NNNSEYRLIN C NTSVITQI CPKVTFDPIP IHYCAPAGYA ILKCNNKTFN GTGPCNNVST VQCTHGIKPV VSTQLLLNGS LAEEEIIIRS EN LTDNVKT IIVHLNESVE ITCTRPNNMT RKSVRIGPGQ TFYALGDIIG DIRQPHCNIS EIKWEKTLQR VSEKLREHFN KTI IFNQSS GGDLEITTHS FNCGGEFFYC NTSDLFFNKT FNETYSTGSN STNSTITLPC RIKQIINMWQ EVGRAMYAPP IAGN ITCKS NITGLLLTRD GGGNNSTKET FRPGGGNMRD NWRSELYKYK VVEVKPLGIA PTECNRTVVQ RRRRRR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #4: 3BNC117 Heavy Chain
| Macromolecule | Name: 3BNC117 Heavy Chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.656484 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLLQSGAA VTKPGASVRV SCEASGYNIR DYFIHWWRQA PGQGLQWVGW INPKTGQPNN PRQFQGRVSL TRHASWDFDT YSFYMDLKA LRSDDTAVYF CARQRSDYWD FDVWGSGTQV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: QVQLLQSGAA VTKPGASVRV SCEASGYNIR DYFIHWWRQA PGQGLQWVGW INPKTGQPNN PRQFQGRVSL TRHASWDFDT YSFYMDLKA LRSDDTAVYF CARQRSDYWD FDVWGSGTQV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSC |
-Macromolecule #5: 3BNC117 Light Chain
| Macromolecule | Name: 3BNC117 Light Chain / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.022658 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDTVT ITCQANGYLN WYQQRRGKAP KLLIYDGSKL ERGVPSRFSG RRWGQEYNLT INNLQPEDIA TYFCQVYEF VVPGTRLDLK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG NSQESVTEQD S KDSTYSLS ...String: DIQMTQSPSS LSASVGDTVT ITCQANGYLN WYQQRRGKAP KLLIYDGSKL ERGVPSRFSG RRWGQEYNLT INNLQPEDIA TYFCQVYEF VVPGTRLDLK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG NSQESVTEQD S KDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Macromolecule #6: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 6 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.355703 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVVGLGAVFL GFLGAAGSTM GAASNTLTVQ ARQLLSGIVQ QQSNLLRAPE AQQHMLQLGV WGFKQLQARV LAIERYLEVQ QLLGMWGCS GKLICCTNVP WNSSWSNKTY NEIWDNMTWM QWDREIGNYT DTIYKLLEVS QFQQEINEKD NLTLD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #11: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 11 / Number of copies: 54 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Formula: PBS |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE |
| Details | PBS |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 66.88 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.91 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 2.15) / Number images used: 35406 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7ly9: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)