[English] 日本語
 Yorodumi
Yorodumi- EMDB-23508: Cryo-EM structure of 13S proteasome core particle assembly interm... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23508 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of 13S proteasome core particle assembly intermediate purified from Pre3-1 proteasome mutant (G34D) | |||||||||
 Map data Map data | Map of 13S complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | core particle / complex / assembly intermediate / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of proteasome core complex assembly / proteasome core complex assembly / nuclear outer membrane-endoplasmic reticulum membrane network / Cross-presentation of soluble exogenous antigens (endosomes) / TNFR2 non-canonical NF-kB pathway / proteasomal ubiquitin-independent protein catabolic process / Ubiquitin Mediated Degradation of Phosphorylated Cdc25A / Regulation of PTEN stability and activity / CDK-mediated phosphorylation and removal of Cdc6 / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis ...regulation of proteasome core complex assembly / proteasome core complex assembly / nuclear outer membrane-endoplasmic reticulum membrane network / Cross-presentation of soluble exogenous antigens (endosomes) / TNFR2 non-canonical NF-kB pathway / proteasomal ubiquitin-independent protein catabolic process / Ubiquitin Mediated Degradation of Phosphorylated Cdc25A / Regulation of PTEN stability and activity / CDK-mediated phosphorylation and removal of Cdc6 / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / KEAP1-NFE2L2 pathway / Neddylation / Orc1 removal from chromatin / MAPK6/MAPK4 signaling / proteasome storage granule / Antigen processing: Ubiquitination & Proteasome degradation / endopeptidase activator activity / proteasome assembly / proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / proteasome core complex, alpha-subunit complex / Ub-specific processing proteases / threonine-type endopeptidase activity / Neutrophil degranulation / proteasome complex / ubiquitin-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process / endopeptidase activity / mRNA binding / DNA damage response / endoplasmic reticulum membrane / mitochondrion / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
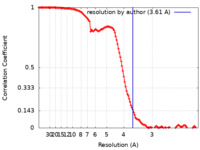
| Method | single particle reconstruction / cryo EM / Resolution: 3.61 Å | |||||||||
 Authors Authors | Schnell HM / Walsh Jr RM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2021 Journal: Nat Struct Mol Biol / Year: 2021Title: Structures of chaperone-associated assembly intermediates reveal coordinated mechanisms of proteasome biogenesis. Authors: Helena M Schnell / Richard M Walsh / Shaun Rawson / Mandeep Kaur / Meera K Bhanu / Geng Tian / Miguel A Prado / Angel Guerra-Moreno / Joao A Paulo / Steven P Gygi / Jeroen Roelofs / Daniel Finley / John Hanna /  Abstract: The proteasome mediates most selective protein degradation. Proteolysis occurs within the 20S core particle (CP), a barrel-shaped chamber with an αββα configuration. CP biogenesis proceeds ...The proteasome mediates most selective protein degradation. Proteolysis occurs within the 20S core particle (CP), a barrel-shaped chamber with an αββα configuration. CP biogenesis proceeds through an ordered multistep pathway requiring five chaperones, Pba1-4 and Ump1. Using Saccharomyces cerevisiae, we report high-resolution structures of CP assembly intermediates by cryogenic-electron microscopy. The first structure corresponds to the 13S particle, which consists of a complete α-ring, partial β-ring (β2-4), Ump1 and Pba1/2. The second structure contains two additional subunits (β5-6) and represents a later pre-15S intermediate. These structures reveal the architecture and positions of Ump1 and β2/β5 propeptides, with important implications for their functions. Unexpectedly, Pba1's N terminus extends through an open CP pore, accessing the CP interior to contact Ump1 and the β5 propeptide. These results reveal how the coordinated activity of Ump1, Pba1 and the active site propeptides orchestrate key aspects of CP assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23508.map.gz emd_23508.map.gz | 168.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23508-v30.xml emd-23508-v30.xml emd-23508.xml emd-23508.xml | 31.5 KB 31.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23508_fsc.xml emd_23508_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_23508.png emd_23508.png | 241.7 KB | ||
| Filedesc metadata |  emd-23508.cif.gz emd-23508.cif.gz | 8.4 KB | ||
| Others |  emd_23508_half_map_1.map.gz emd_23508_half_map_1.map.gz emd_23508_half_map_2.map.gz emd_23508_half_map_2.map.gz | 164.9 MB 164.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23508 http://ftp.pdbj.org/pub/emdb/structures/EMD-23508 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23508 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23508 | HTTPS FTP |
-Validation report
| Summary document |  emd_23508_validation.pdf.gz emd_23508_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23508_full_validation.pdf.gz emd_23508_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_23508_validation.xml.gz emd_23508_validation.xml.gz | 20.8 KB | Display | |
| Data in CIF |  emd_23508_validation.cif.gz emd_23508_validation.cif.gz | 26.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23508 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23508 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23508 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23508 | HTTPS FTP |
-Related structure data
| Related structure data |  7lsxMC  7ls5C  7ls6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23508.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23508.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of 13S complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
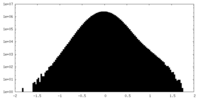
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half Map 1 for 13S Complex
| File | emd_23508_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 for 13S Complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
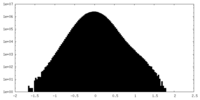
| Density Histograms |
-Half map: Half Map 2 for 13S Complex
| File | emd_23508_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 for 13S Complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
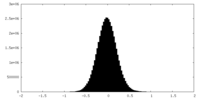
| Density Histograms |
- Sample components
Sample components
+Entire : 13S
+Supramolecule #1: 13S
+Macromolecule #1: Proteasome subunit alpha type-1
+Macromolecule #2: Proteasome subunit alpha type-2
+Macromolecule #3: Proteasome subunit alpha type-3
+Macromolecule #4: Proteasome subunit alpha type-4
+Macromolecule #5: Proteasome subunit alpha type-5
+Macromolecule #6: Proteasome subunit alpha type-6
+Macromolecule #7: Proteasome subunit alpha type-7
+Macromolecule #8: Proteasome maturation factor UMP1
+Macromolecule #9: Proteasome subunit beta type-2
+Macromolecule #10: Proteasome subunit beta type-3
+Macromolecule #11: Proteasome subunit beta type-4
+Macromolecule #12: Proteasome chaperone 1
+Macromolecule #13: Proteasome assembly chaperone 2
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Fluorinated Fos-Choline was added to the sample immediately prior to deposition on a grid for plunge freezing. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 25 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 20657 / Average exposure time: 2.4 sec. / Average electron dose: 55.94 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 47169 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)