+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23336 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Phage Qbeta virion | |||||||||||||||
 Map data Map data | Qbeta Virion | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsuppression by virus of host cell wall biogenesis / suppression by virus of host peptidoglycan biosynthetic process / viral release via suppression of host peptidoglycan biosynthetic process / virion attachment to host cell pilus / T=3 icosahedral viral capsid / adhesion receptor-mediated virion attachment to host cell / translation repressor activity / viral release from host cell by cytolysis / virion component / structural molecule activity / RNA binding Similarity search - Function | |||||||||||||||
| Biological species |  Escherichia virus Qbeta / Escherichia virus Qbeta /  Bacteriophage Q-beta (virus) Bacteriophage Q-beta (virus) | |||||||||||||||
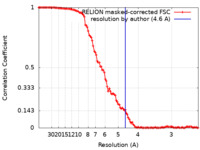
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||||||||
 Authors Authors | Chang JY / Zhang J | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Viruses / Year: 2022 Journal: Viruses / Year: 2022Title: Structural Assembly of Qβ Virion and Its Diverse Forms of Virus-like Particles. Authors: Jeng-Yih Chang / Karl V Gorzelnik / Jirapat Thongchol / Junjie Zhang /  Abstract: The coat proteins (CPs) of single-stranded RNA bacteriophages (ssRNA phages) directly assemble around the genomic RNA (gRNA) to form a near-icosahedral capsid with a single maturation protein (Mat) ...The coat proteins (CPs) of single-stranded RNA bacteriophages (ssRNA phages) directly assemble around the genomic RNA (gRNA) to form a near-icosahedral capsid with a single maturation protein (Mat) that binds the gRNA and interacts with the retractile pilus during infection of the host. Understanding the assembly of ssRNA phages is essential for their use in biotechnology, such as RNA protection and delivery. Here, we present the complete gRNA model of the ssRNA phage Qβ, revealing that the 3' untranslated region binds to the Mat and the 4127 nucleotides fold domain-by-domain, and is connected through long-range RNA-RNA interactions, such as kissing loops. Thirty-three operator-like RNA stem-loops are located and primarily interact with the asymmetric A/B CP-dimers, suggesting a pathway for the assembly of the virions. Additionally, we have discovered various forms of the virus-like particles (VLPs), including the canonical = 3 icosahedral, larger = 4 icosahedral, prolate, oblate forms, and a small prolate form elongated along the 3-fold axis. These particles are all produced during a normal infection, as well as when overexpressing the CPs. When overexpressing the shorter RNA fragments encoding only the CPs, we observed an increased percentage of the smaller VLPs, which may be sufficient to encapsidate a shorter RNA. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23336.map.gz emd_23336.map.gz | 22.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23336-v30.xml emd-23336-v30.xml emd-23336.xml emd-23336.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23336_fsc.xml emd_23336_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_23336.png emd_23336.png | 273.7 KB | ||
| Others |  emd_23336_additional_1.map.gz emd_23336_additional_1.map.gz | 97.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23336 http://ftp.pdbj.org/pub/emdb/structures/EMD-23336 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23336 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23336 | HTTPS FTP |
-Validation report
| Summary document |  emd_23336_validation.pdf.gz emd_23336_validation.pdf.gz | 464.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23336_full_validation.pdf.gz emd_23336_full_validation.pdf.gz | 464.4 KB | Display | |
| Data in XML |  emd_23336_validation.xml.gz emd_23336_validation.xml.gz | 10.4 KB | Display | |
| Data in CIF |  emd_23336_validation.cif.gz emd_23336_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23336 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23336 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23336 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23336 | HTTPS FTP |
-Related structure data
| Related structure data |  7lhdMC  7lgeC  7lgfC  7lggC  7lghC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23336.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23336.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
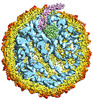
| Annotation | Qbeta Virion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.25 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened Map
| File | emd_23336_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened Map | ||||||||||||
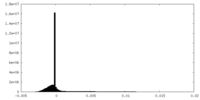
| Projections & Slices |
| ||||||||||||
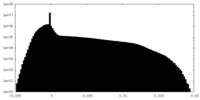
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia virus Qbeta
| Entire | Name:  Escherichia virus Qbeta Escherichia virus Qbeta |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia virus Qbeta
| Supramolecule | Name: Escherichia virus Qbeta / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 39803 / Sci species name: Escherichia virus Qbeta / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Genomic RNA
| Macromolecule | Name: Genomic RNA / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Escherichia virus Qbeta Escherichia virus Qbeta |
| Molecular weight | Theoretical: 1.3520166249999999 MDa |
| Sequence | String: GGGGACCCCC UUUAGGGGGU CACCUCACAC AGCAGUACUU CACUGAGUAU AAGAGGACAU AUGCCUAAAU UACCGCGUGG UCUGCGUUU CGGAGCCGAU AAUGAAAUUC UUAAUGAUUU UCAGGAGCUC UGGUUUCCAG ACCUCUUUAU CGAAUCUUCC G ACACGCAU ...String: GGGGACCCCC UUUAGGGGGU CACCUCACAC AGCAGUACUU CACUGAGUAU AAGAGGACAU AUGCCUAAAU UACCGCGUGG UCUGCGUUU CGGAGCCGAU AAUGAAAUUC UUAAUGAUUU UCAGGAGCUC UGGUUUCCAG ACCUCUUUAU CGAAUCUUCC G ACACGCAU CCGUGGUACA CACUGAAGGG UCGUGUGUUG AACGCCCACC UUGAUGAUCG UCUACCUAAU GUAGGCGGUC GC CAGGUAA GGCGCACUCC ACAUCGCGUC ACCGUUCCGA UUGCCUCUUC AGGCCUUCGU CCGGUAACAA CCGUUCAGUA UGA UCCCGC AGCACUAUCG UUCUUAUUGA ACGCUCGUGU UGACUGGGAU UUCGGUAAUG GCGAUAGUGC GAACCUUGUC AUUA AUGAC UUUCUGUUUC GCACCUUUGC ACCUAAGGAG UUUGAUUUUU CGAACUCCUU AGUUCCUCGU UAUACUCAGG CCUUC UCCG CGUUUAAUGC CAAGUAUGGC ACUAUGAUCG GCGAAGGGCU CGAGACUAUA AAAUAUCUCG GGCUUUUACU GCGCAG ACU GCGUGAGGGU UACCGCGCUG UUAAGCGUGG CGAUUUACGU GCUCUUCGUA GGGUUAUCCA GUCCUACCAU AAUGGUA AG UGGAAACCGG CUACUGCUGG UAAUCUCUGG CUUGAAUUUC GUUAUGGCCU UAUGCCUCUC UUUUAUGACA UCAGAGAU G UCAUGUUAGA CUGGCAGAAC CGUCAUGAUA AGAUUCAACG CCUCCUUCGG UUUUCUGUUG GUCACGGCGA GGAUUACGU UGUCGAAUUC GACAAUCUGU ACCCUGCCGU UGCUUACUUU AAACUGAAAG GGGAGAUUAC ACUCGAACGC CGUCAUCGUC AUGGCAUAU CUUACGCUAA CCGCGAAGGA UAUGCUGUUU UCGACAACGG UUCCCUUCGG CCUGUGUCCG AUUGGAAGGA G CUUGCCAC UGCAUUCAUC AAUCCGCAUG AAGUUGCUUG GGAGUUAACU CCCUACAGCU UCGUUGUUGA UUGGUUCUUG AA UGUUGGU GACAUACUUG CUCAACAAGG UCAGCUAUAU CAUAAUAUCG AUAUUGUAGA CGGCUUUGAC AGACGUGACA UCC GGCUCA AAUCUUUCAC CAUAAAAGGU GAACGAAAUG GGCGGCCUGU UAACGUUUCU GCUAGCCUGU CUGCUGUCGA UUUA UUUUA CAGCCGACUC CAUACGAGCA AUCUUCCGUU CGCUACACUA GAUCUUGAUA CCACCUUUAG UUCGUUUAAA CACGU UCUU GAUAGUAUCU UUUUAUUAAC CCAACGCGUA AAGCGUUGAA ACUUUGGGUC AAUUUGAUCA UGGCAAAAUU AGAGAC UGU UACUUUAGGU AACAUCGGGA AAGAUGGAAA ACAAACUCUG GUCCUCAAUC CGCGUGGGGU AAAUCCCACU AACGGCG UU GCCUCGCUUU CACAAGCGGG UGCAGUUCCU GCGCUGGAGA AGCGUGUUAC CGUUUCGGUA UCUCAGCCUU CUCGCAAU C GUAAGAACUA CAAGGUCCAG GUUAAGAUCC AGAACCCGAC CGCUUGCACU GCAAACGGUU CUUGUGACCC AUCCGUUAC UCGCCAGGCA UAUGCUGACG UGACCUUUUC GUUCACGCAG UAUAGUACCG AUGAGGAACG AGCUUUUGUU CGUACAGAGC UUGCUGCUC UGCUCGCUAG UCCUCUGCUG AUCGAUGCUA UUGAUCAGCU GAACCCAGCG UAUUGAACAC UGCUCAUUGC C GGUGGUGG CUCAGGGUCA AAACCCGAUC CGGUUAUUCC GGAUCCACCG AUUGAUCCGC CGCCAGGGAC AGGUAAGUAU AC CUGUCCC UUCGCAAUUU GGUCCCUAGA GGAGGUUUAC GAGCCUCCUA CUAAGAACCG ACCGUGGCCU AUCUAUAAUG CUG UUGAAC UCCAGCCUCG CGAAUUUGAU GUUGCCCUCA AAGAUCUUUU GGGCAAUACA AAGUGGCGUG AUUGGGAUUC UCGG CUUAG UUAUACCACG UUCCGCGGUU GCCGUGGCAA UGGUUAUAUU GACCUUGAUG CGACUUAUCU UGCUACUGAU CAGGC UAUG CGUGAUCAGA AGUAUGAUAU UCGCGAGGGC AAGAAACCUG GUGCUUUCGG UAACAUUGAG CGAUUCAUUU AUCUUA AGU CGAUAAAUGC UUAUUGCUCU CUUAGCGAUA UUGCGGCCUA UCACGCCGAU GGCGUGAUAG UUGGCUUUUG GCGCGAU CC AUCCAGCGGU GGUGCCAUAC CGUUUGACUU CACUAAGUUU GAUAAGACUA AAUGUCCUAU UCAAGCCGUG AUAGUCGU U CCUCGUGCUU AGUAACUAAG GAUGAAAUGC AUGUCUAAGA CAGCAUCUUC GCGUAACUCU CUCAGCGCAC AAUUGCGCC GAGCCGCGAA CACAAGAAUU GAGGUUGAAG GUAACCUCGC ACUUUCCAUU GCCAACGAUU UACUGUUGGC CUAUGGUCAG UCGCCAUUU AACUCUGAGG CUGAGUGUAU UUCAUUCAGC CCGAGAUUCG ACGGGACCCC GGAUGACUUU AGGAUAAAUU A UCUUAAAG CCGAGAUCAU GUCGAAGUAU GACGACUUCA GCCUAGGUAU UGAUACCGAA GCUGUUGCCU GGGAGAAGUU CC UGGCAGC AGAGGCUGAA UGUGCUUUAA CGAACGCUCG UCUCUAUAGG CCUGACUACA GUGAGGAUUU CAAUUUCUCA CUG GGCGAG UCAUGUAUAC ACAUGGCUCG UAGAAAAAUA GCCAAGCUAA UAGGAGAUGU UCCGUCCGUU GAGGGUAUGU UGCG UCACU GCCGAUUUUC UGGCGGUGCU ACAACAACGA AUAACCGUUC GUACGGUCAU CCGUCCUUCA AGUUUGCGCU UCCGC AAGC GUGUACGCCU CGGGCUUUGA AGUAUGUUUU AGCUCUCAGA GCUUCUACAC AUUUCGAUAU CAGAAUUUCU GAUAUU AGC CCUUUUAAUA AAGCAGUUAC UGUACCUAAG AACAGUAAGA CAGAUCGUUG UAUUGCUAUC GAACCUGGUU GGAAUAU GU UUUUCCAACU GGGUAUCGGU GGCAUUCUAC GCGAUCGGUU GCGUUGCUGG GGUAUCGAUC UGAAUGAUCA GACGAUAA A UCAGCGCCGC GCUCACGAAG GCUCCGUUAC UAAUAACUUA GCAACGGUUG AUCUCUCAGC GGCAAGCGAU UCUAUAUCU CUUGCCCUCU GUGAGCUCUU AUUGCCCCCA GGCUGGUUUG AGGUUCUUAU GGACCUCAGA UCACCUAAGG GGCGAUUGCC UGACGGUAG UGUUGUUACC UACGAGAAGA UUUCUUCUAU GGGUAACGGU UACACAUUCG AGCUCGAGUC GCUUAUUUUU G CUUCUCUC GCUCGUUCCG UUUGUGAGAU ACUGGACUUA GACUCGUCUG AGGUCACUGU UUACGGAGAC GAUAUUAUUU UA CCGUCCU GUGCAGUCCC UGCCCUCCGG GAAGUUUUUA AGUAUGUUGG UUUUACGACC AAUACUAAAA AGACUUUUUC CGA GGGGCC GUUCAGAGAG UCGUGCGGCA AGCACUACUA UUCUGGCGUA GAUGUUACUC CCUUUUACAU ACGUCACCGU AUAG UGAGU CCUGCCGAUU UAAUACUGGU UUUGAAUAAC CUAUAUCGGU GGGCCACAAU UGACGGCGUA UGGGAUCCUA GGGCC CAUU CUGUGUACCU CAAGUAUCGU AAGUUGCUGC CUAAACAGCU GCAACGUAAU ACUAUACCUG AUGGUUACGG UGAUGG UGC CCUCGUCGGA UCGGUCCUAA UCAAUCCUUU CGCGAAAAAC CGCGGGUGGA UCCGGUACGU ACCGGUGAUU ACGGACC AU ACAAGGGACC GAGAGCGCGC UGAGUUGGGG UCGUAUCUCU ACGACCUCUU CUCGCGUUGU CUCUCGGAAA GUAACGAU G GGUUGCCUCU UAGGGGUCCA UCGGGUUGCG AUUCUGCGGA UCUAUUUGCC AUCGAUCAGC UUAUCUGUAG GAGUAAUCC UACGAAGAUA AGCAGGUCUA CCGGCAAAUU CGAUAUACAG UAUAUCGCGU GCAGUAGCCG UGUUCUGGCA CCCUACGGGG UCUUCCAGG GCACGAAGGU UGCGUCUCUA CACGAGGCGU AACCUGGGAG GGCGCCAAUA UGGCGCCUAA UUGUGAAUAA A UUAUCACA AUUACUCUUA CGAGUGAGAG GGGGAUCUGC UUUGCCCUCU CUCCUCCCA |
-Macromolecule #2: Maturation protein A2
| Macromolecule | Name: Maturation protein A2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteriophage Q-beta (virus) Bacteriophage Q-beta (virus) |
| Molecular weight | Theoretical: 48.613078 KDa |
| Sequence | String: MPKLPRGLRF GADNEILNDF QELWFPDLFI ESSDTHPWYT LKGRVLNAHL DDRLPNVGGR QVRRTPHRVT VPIASSGLRP VTTVQYDPA ALSFLLNARV DWDFGNGDSA NLVINDFLFR TFAPKEFDFS NSLVPRYTQA FSAFNAKYGT MIGEGLETIK Y LGLLLRRL ...String: MPKLPRGLRF GADNEILNDF QELWFPDLFI ESSDTHPWYT LKGRVLNAHL DDRLPNVGGR QVRRTPHRVT VPIASSGLRP VTTVQYDPA ALSFLLNARV DWDFGNGDSA NLVINDFLFR TFAPKEFDFS NSLVPRYTQA FSAFNAKYGT MIGEGLETIK Y LGLLLRRL REGYRAVKRG DLRALRRVIQ SYHNGKWKPA TAGNLWLEFR YGLMPLFYDI RDVMLDWQNR HDKIQRLLRF SV GHGEDYV VEFDNLYPAV AYFKLKGEIT LERRHRHGIS YANREGYAVF DNGSLRPVSD WKELATAFIN PHEVAWELTP YSF VVDWFL NVGDILAQQG QLYHNIDIVD GFDRRDIRLK SFTIKGERNG RPVNVSASLS AVDLFYSRLH TSNLPFATLD LDTT FSSFK HVLDSIFLLT QRVKR |
-Macromolecule #3: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 3 / Number of copies: 180 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteriophage Q-beta (virus) Bacteriophage Q-beta (virus) |
| Molecular weight | Theoretical: 14.268071 KDa |
| Sequence | String: MAKLETVTLG NIGKDGKQTL VLNPRGVNPT NGVASLSQAG AVPALEKRVT VSVSQPSRNR KNYKVQVKIQ NPTACTANGS CDPSVTRQA YADVTFSFTQ YSTDEERAFV RTELAALLAS PLLIDAIDQL NPAY |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | JEOL 3200FSC |
| Image recording | Image recording ID: 1 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TECNAI F20 |
| Image recording | Image recording ID: 2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z
Z Y
Y X
X