+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22819 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | HCMV postfusion gB in complex with SM5-1 Fab | |||||||||
 Map data Map data | HCMV postfusion gB in complex with SM5-1 FAb | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / host cell endosome membrane / symbiont entry into host cell / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Human cytomegalovirus (strain Towne) / Human cytomegalovirus (strain Towne) /  Homo sapiens (human) / Homo sapiens (human) /   HHV-5 (virus) HHV-5 (virus) | |||||||||
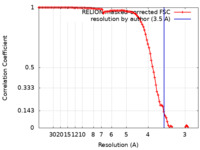
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Liu Y / Heim PK / Che Y / Chi X / Qiu X / Han S / Dormitzer PR / Yang X | |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion. Authors: Yuhang Liu / Kyle P Heim / Ye Che / Xiaoyuan Chi / Xiayang Qiu / Seungil Han / Philip R Dormitzer / Xinzhen Yang /  Abstract: Human cytomegalovirus (HCMV) causes congenital disease with long-term morbidity. HCMV glycoprotein B (gB) transitions irreversibly from a metastable prefusion to a stable postfusion conformation to ...Human cytomegalovirus (HCMV) causes congenital disease with long-term morbidity. HCMV glycoprotein B (gB) transitions irreversibly from a metastable prefusion to a stable postfusion conformation to fuse the viral envelope with a host cell membrane during entry. We stabilized prefusion gB on the virion with a fusion inhibitor and a chemical cross-linker, extracted and purified it, and then determined its structure to 3.6-Å resolution by electron cryomicroscopy. Our results revealed the structural rearrangements that mediate membrane fusion and details of the interactions among the fusion loops, the membrane-proximal region, transmembrane domain, and bound fusion inhibitor that stabilized gB in the prefusion state. The structure rationalizes known gB antigenic sites. By analogy to successful vaccine antigen engineering approaches for other viral pathogens, the high-resolution prefusion gB structure provides a basis to develop stabilized prefusion gB HCMV vaccine antigens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22819.map.gz emd_22819.map.gz | 5.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22819-v30.xml emd-22819-v30.xml emd-22819.xml emd-22819.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22819_fsc.xml emd_22819_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_22819.png emd_22819.png | 44.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22819 http://ftp.pdbj.org/pub/emdb/structures/EMD-22819 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22819 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22819 | HTTPS FTP |
-Validation report
| Summary document |  emd_22819_validation.pdf.gz emd_22819_validation.pdf.gz | 353.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22819_full_validation.pdf.gz emd_22819_full_validation.pdf.gz | 352.7 KB | Display | |
| Data in XML |  emd_22819_validation.xml.gz emd_22819_validation.xml.gz | 11.5 KB | Display | |
| Data in CIF |  emd_22819_validation.cif.gz emd_22819_validation.cif.gz | 15.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22819 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22819 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22819 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22819 | HTTPS FTP |
-Related structure data
| Related structure data |  7kddMC  7kdpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22819.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22819.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HCMV postfusion gB in complex with SM5-1 FAb | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.376 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HCMV postfusion gB in complex with a neutralizing antibody Fab SM5-1
| Entire | Name: HCMV postfusion gB in complex with a neutralizing antibody Fab SM5-1 |
|---|---|
| Components |
|
-Supramolecule #1: HCMV postfusion gB in complex with a neutralizing antibody Fab SM5-1
| Supramolecule | Name: HCMV postfusion gB in complex with a neutralizing antibody Fab SM5-1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: SM5-1 FAb is recombinant expressed with 6xHis and Strep sequence at C-terminal as purification tags. The FAb was used to purify the gB protein from lab cultured HCMV virus (Towne strain) |
|---|---|
| Molecular weight | Theoretical: 300 KDa |
-Supramolecule #2: Envelope glycoprotein B
| Supramolecule | Name: Envelope glycoprotein B / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Human cytomegalovirus (strain Towne) / Strain: Towne Human cytomegalovirus (strain Towne) / Strain: Towne |
-Supramolecule #3: SM5-1 Fab antibody heavy chain, SM5-1 Fab antibody light chain
| Supramolecule | Name: SM5-1 Fab antibody heavy chain, SM5-1 Fab antibody light chain type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Scombridae gen. sp. (tuna) Scombridae gen. sp. (tuna) |
-Macromolecule #1: Envelope glycoprotein B
| Macromolecule | Name: Envelope glycoprotein B / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   HHV-5 (virus) / Strain: Towne HHV-5 (virus) / Strain: Towne |
| Molecular weight | Theoretical: 102.067688 KDa |
| Sequence | String: MESRIWCLVV CVNLCIVCLG AAVSSSSTRG TSATHSHHSS HTTSAAHSRS GSVSQRVTSS QTVSHGVNET IYNTTLKYGD VVGVNTTKY PYRVCSMAQG TDLIRFERNI VCTSMKPINE DLDEGIMVVY KRNIVAHTFK VRVYQKVLTF RRSYAYIHTT Y LLGSNTEY ...String: MESRIWCLVV CVNLCIVCLG AAVSSSSTRG TSATHSHHSS HTTSAAHSRS GSVSQRVTSS QTVSHGVNET IYNTTLKYGD VVGVNTTKY PYRVCSMAQG TDLIRFERNI VCTSMKPINE DLDEGIMVVY KRNIVAHTFK VRVYQKVLTF RRSYAYIHTT Y LLGSNTEY VAPPMWEIHH INSHSQCYSS YSRVIAGTVF VAYHRDSYEN KTMQLMPDDY SNTHSTRYVT VKDQWHSRGS TW LYRETCN LNCMVTITTA RSKYPYHFFA TSTGDVVDIS PFYNGTNRNA SYFGENADKF FIFPNYTIVS DFGRPNSALE THR LVAFLE RADSVISWDI QDEKNVTCQL TFWEASERTI RSEAEDSYHF SSAKMTATFL SKKQEVNMSD SALDCVRDEA INKL QQIFN TSYNQTYEKY GNVSVFETTG GLVVFWQGIK QKSLVELERL ANRSSLNLTH NRTKRSTDGN NATHLSNMES VHNLV YAQL QFTYDTLRGY INRALAQIAE AWCVDQRRTL EVFKELSKIN PSAILSAIYN KPIAARFMGD VLGLASCVTI NQTSVK VLR DMNVKESPGR CYSRPVVIFN FANSSYVQYG QLGEDNEILL GNHRTEECQL PSLKIFIAGN SAYEYVDYLF KRMIDLS SI STVDSMIALD IDPLENTDFR VLELYSQKEL RSSNVFDLEE IMREFNSYKQ RVKYVEDKVV DPLPPYLKGL DDLMSGLG A AGKAVGVAIG AVGGAVASVV EGVATFLKNP FGAFTIILVA IAVVIIIYLI YTRQRRLCMQ PLQNLFPYLV SADGTTVTS GNTKDTSLQA PPSYEESVYN SGRKGPGPPS SDASTAAPPY TNEQAYQMLL ALVRLDAEQR AQQNGTDSLD GQTGTQDKGQ KPNLLDRLR HRKNGYRHLK DSDEEENV |
-Macromolecule #2: SM5-1 Fab antibody heavy chain
| Macromolecule | Name: SM5-1 Fab antibody heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.534768 KDa |
| Recombinant expression | Organism:  Scombridae gen. sp. (tuna) Scombridae gen. sp. (tuna) |
| Sequence | String: QVQLVQSGAE VRKPGASVKV SCKASGYSLK DHYMVWVRQA PGQGLEWMGW INPQSGGTGY GQKFQGRVTM TRDTSTNTAY MILSSLRSD DTAVYFCARD GAKTVSNSGL SLLYYHNRLD AWGQGTMVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL V KDYFPEPV ...String: QVQLVQSGAE VRKPGASVKV SCKASGYSLK DHYMVWVRQA PGQGLEWMGW INPQSGGTGY GQKFQGRVTM TRDTSTNTAY MILSSLRSD DTAVYFCARD GAKTVSNSGL SLLYYHNRLD AWGQGTMVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL V KDYFPEPV TVSWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCGGGG GA GGGGGHH HHHHGSWSHP QFEK |
-Macromolecule #3: SM5-1 Fab antibody light chain
| Macromolecule | Name: SM5-1 Fab antibody light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: Towne Homo sapiens (human) / Strain: Towne |
| Molecular weight | Theoretical: 22.638217 KDa |
| Recombinant expression | Organism:  Scombridae gen. sp. (tuna) Scombridae gen. sp. (tuna) |
| Sequence | String: QSVLTQPPSV SAAPGQMVTI SCSGSSSNIG KNYVSWYQQL PGAAPKLLIF DNNKRPSGTP DRFSGSKSGT SATLVITGLQ TGDEADYYC GTPDRSLSVI FGGGTKVTVL GQPKANPTVT LFPPSSEELQ ANKATLVCLI SDFYPGAVTV AWKADGSPVK A GVETTKPS ...String: QSVLTQPPSV SAAPGQMVTI SCSGSSSNIG KNYVSWYQQL PGAAPKLLIF DNNKRPSGTP DRFSGSKSGT SATLVITGLQ TGDEADYYC GTPDRSLSVI FGGGTKVTVL GQPKANPTVT LFPPSSEELQ ANKATLVCLI SDFYPGAVTV AWKADGSPVK A GVETTKPS KQSNNKYAAS SYLSLTPEQW KSHRSYSCQV THEGSTVEKT VAPTECS |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 30 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS, 0.1 % DDM, 1 mM EDTA, 2 mg/L WAY-174865 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK II / Details: 4 second, -2 force. |
| Details | this sample was monodispersed |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-28 / Number grids imaged: 3 / Number real images: 7768 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 18000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller