[English] 日本語
 Yorodumi
Yorodumi- EMDB-22513: Satellite phage P4 procapsid including size determination (Sid) p... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22513 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Satellite phage P4 procapsid including size determination (Sid) protein | |||||||||
 Map data Map data | Final masked and sharpened map reconstructed without applying symmetry from symmetry expanded particle. Normalized to mean=0 and 1 stddev. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteriophage / phage / procapsid / satellite phage / size determination / capsid protein / molecular piracy / VIRUS | |||||||||
| Function / homology | Bacteriophage P2, capsid / Phage major capsid protein, P2 family / viral capsid / Glycoprotein 3 / Capsid proteins Function and homology information Function and homology information | |||||||||
| Biological species |   Escherichia phage P2 (virus) / Escherichia phage P2 (virus) /  Enterobacteria phage P4 (virus) Enterobacteria phage P4 (virus) | |||||||||
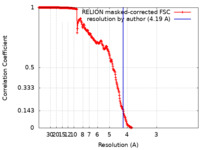
| Method | single particle reconstruction / cryo EM / Resolution: 4.19 Å | |||||||||
 Authors Authors | Kizziah JL / Dokland T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Viruses / Year: 2020 Journal: Viruses / Year: 2020Title: Structure of the Capsid Size-Determining Scaffold of "Satellite" Bacteriophage P4. Authors: James L Kizziah / Cynthia M Rodenburg / Terje Dokland /  Abstract: P4 is a mobile genetic element (MGE) that can exist as a plasmid or integrated into its host genome, but becomes packaged into phage particles by a helper bacteriophage, such as P2. P4 is the ...P4 is a mobile genetic element (MGE) that can exist as a plasmid or integrated into its host genome, but becomes packaged into phage particles by a helper bacteriophage, such as P2. P4 is the original example of what we have termed "molecular piracy", the process by which one MGE usurps the life cycle of another for its own propagation. The P2 helper provides most of the structural gene products for assembly of the P4 virion. However, when P4 is mobilized by P2, the resulting capsids are smaller than those normally formed by P2 alone. The P4-encoded protein responsible for this size change is called Sid, which forms an external scaffolding cage around the P4 procapsids. We have determined the high-resolution structure of P4 procapsids, allowing us to build an atomic model for Sid as well as the gpN capsid protein. Sixty copies of Sid form an intertwined dodecahedral cage around the = 4 procapsid, making contact with only one out of the four symmetrically non-equivalent copies of gpN. Our structure provides a basis for understanding the mutants in gpN that prevent small capsid formation, as well as the "super-sid" mutations that counteract the effect of the mutations, and suggests a model for capsid size redirection by Sid. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22513.map.gz emd_22513.map.gz | 2.3 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22513-v30.xml emd-22513-v30.xml emd-22513.xml emd-22513.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22513_fsc.xml emd_22513_fsc.xml | 30.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_22513.png emd_22513.png | 310.7 KB | ||
| Masks |  emd_22513_msk_1.map emd_22513_msk_1.map | 2.4 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-22513.cif.gz emd-22513.cif.gz | 5.5 KB | ||
| Others |  emd_22513_half_map_1.map.gz emd_22513_half_map_1.map.gz emd_22513_half_map_2.map.gz emd_22513_half_map_2.map.gz | 2 GB 2 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22513 http://ftp.pdbj.org/pub/emdb/structures/EMD-22513 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22513 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22513 | HTTPS FTP |
-Validation report
| Summary document |  emd_22513_validation.pdf.gz emd_22513_validation.pdf.gz | 976.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22513_full_validation.pdf.gz emd_22513_full_validation.pdf.gz | 975.8 KB | Display | |
| Data in XML |  emd_22513_validation.xml.gz emd_22513_validation.xml.gz | 38.1 KB | Display | |
| Data in CIF |  emd_22513_validation.cif.gz emd_22513_validation.cif.gz | 52 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22513 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22513 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22513 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22513 | HTTPS FTP |
-Related structure data
| Related structure data |  7jw1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22513.map.gz / Format: CCP4 / Size: 2.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22513.map.gz / Format: CCP4 / Size: 2.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final masked and sharpened map reconstructed without applying symmetry from symmetry expanded particle. Normalized to mean=0 and 1 stddev. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
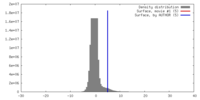
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22513_msk_1.map emd_22513_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
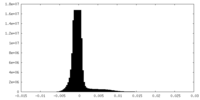
| Density Histograms |
-Half map: Half map 1 from auto-refinement of final map.
| File | emd_22513_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 from auto-refinement of final map. | ||||||||||||
| Projections & Slices |
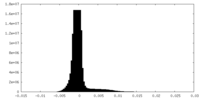
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 from auto-refinement of final map.
| File | emd_22513_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 from auto-refinement of final map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : P4 procapsid
| Entire | Name: P4 procapsid |
|---|---|
| Components |
|
-Supramolecule #1: P4 procapsid
| Supramolecule | Name: P4 procapsid / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Major capsid protein gpN
| Macromolecule | Name: Major capsid protein gpN / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage P2 (virus) Escherichia phage P2 (virus) |
| Molecular weight | Theoretical: 40.291484 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRQETRFKFN AYLSRVAELN GIDAGDVSKK FTVEPSVTQT LMNTMQESSD FLTRINIVPV SEMKGEKIGI GVTGSIASTT DTAGGTERQ PKDFSKLASN KYECDQINFD FYIRYKTLDL WARYQDFQLR IRNAIIKRQS LDFIMAGFNG VKRAETSDRS S NPMLQDVA ...String: MRQETRFKFN AYLSRVAELN GIDAGDVSKK FTVEPSVTQT LMNTMQESSD FLTRINIVPV SEMKGEKIGI GVTGSIASTT DTAGGTERQ PKDFSKLASN KYECDQINFD FYIRYKTLDL WARYQDFQLR IRNAIIKRQS LDFIMAGFNG VKRAETSDRS S NPMLQDVA VGWLQKYRNE APARVMSKVT DEEGRTTSEV IRVGKGGDYA SLDALVMDAT NNLIEPWYQE DPDLVVIVGR QL LADKYFP IVNKEQDNSE MLAADVIISQ KRIGNLPAVR VPYFPADAML ITKLENLSIY YMDDSHRRVI EENPKLDRVE NYE SMNIDY VVEDYAAGCL VEKIKVGDFS TPAKATAEPG A UniProtKB: Capsid proteins |
-Macromolecule #2: Size determination protein Sid
| Macromolecule | Name: Size determination protein Sid / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage P4 (virus) Enterobacteria phage P4 (virus) |
| Molecular weight | Theoretical: 27.296814 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSDHTIPEYL QPALAQLEKA RAAHLENARL MDETVTAIER AEQEKNALAQ ADGNDADDWR TAFRAAGGVL SDELKQRHIE RVARRELVQ EYDNLAVVLN FERERLKGAC DSTATAYRKA HHHLLSLYAE HELEHALNET CEALVRAMHL SILVQENPLA N TTGHQGYV ...String: MSDHTIPEYL QPALAQLEKA RAAHLENARL MDETVTAIER AEQEKNALAQ ADGNDADDWR TAFRAAGGVL SDELKQRHIE RVARRELVQ EYDNLAVVLN FERERLKGAC DSTATAYRKA HHHLLSLYAE HELEHALNET CEALVRAMHL SILVQENPLA N TTGHQGYV APEKAVMQQV KSSLEQKIKQ MQISLTGEPV LRLTGLSAAT LPHMDYEVAG TPAQRKVWQD KIDQQGAELK AR GLLS UniProtKB: Glycoprotein 3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)