+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22333 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SPN3US phage mottled capsid | |||||||||
 Map data Map data | SPN3US mottled capsid | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Salmonella phage SPN3US (virus) Salmonella phage SPN3US (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Heymann JB / Wang B / Newcomb WW / Wu W / Winkler DC / Cheng N / Reilly ER / Hsia R-C / Thomas JA / Steven AC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Viruses / Year: 2020 Journal: Viruses / Year: 2020Title: The Mottled Capsid of the Giant Phage SPN3US, a Likely Maturation Intermediate with a Novel Internal Shell. Authors: J Bernard Heymann / Bing Wang / William W Newcomb / Weimin Wu / Dennis C Winkler / Naiqian Cheng / Erin R Reilly / Ru-Ching Hsia / Julie A Thomas / Alasdair C Steven /  Abstract: "Giant" phages have genomes of >200 kbp, confined in correspondingly large capsids whose assembly and maturation are still poorly understood. Nevertheless, the first assembly product is likely to be, ..."Giant" phages have genomes of >200 kbp, confined in correspondingly large capsids whose assembly and maturation are still poorly understood. Nevertheless, the first assembly product is likely to be, as in other tailed phages, a procapsid that subsequently matures and packages the DNA. The associated transformations include the cleavage of many proteins by the phage-encoded protease, as well as the thinning and angularization of the capsid. We exploited an amber mutation in the viral protease gene of the giant phage SPN3US, which leads to the accumulation of a population of capsids with distinctive properties. Cryo-electron micrographs reveal patterns of internal density different from those of the DNA-filled heads of virions, leading us to call them "mottled capsids". Reconstructions show an outer shell with T = 27 symmetry, an embellishment of the HK97 prototype composed of the major capsid protein, gp75, which is similar to some other giant viruses. The mottled capsid has a T = 1 inner icosahedral shell that is a complex network of loosely connected densities composed mainly of the ejection proteins gp53 and gp54. Segmentation of this inner shell indicated that a number of densities (~12 per asymmetric unit) adopt a "twisted hook" conformation. Large patches of a proteinaceous tetragonal lattice with a 67 Å repeat were also present in the cell lysate. The unexpected nature of these novel inner shell and lattice structures poses questions as to their functions in virion assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22333.map.gz emd_22333.map.gz | 441.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22333-v30.xml emd-22333-v30.xml emd-22333.xml emd-22333.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22333_fsc.xml emd_22333_fsc.xml | 17.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_22333.png emd_22333.png | 215.7 KB | ||
| Masks |  emd_22333_msk_1.map emd_22333_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Others |  emd_22333_additional.map.gz emd_22333_additional.map.gz emd_22333_half_map_1.map.gz emd_22333_half_map_1.map.gz emd_22333_half_map_2.map.gz emd_22333_half_map_2.map.gz | 193.1 MB 442.3 MB 442.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22333 http://ftp.pdbj.org/pub/emdb/structures/EMD-22333 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22333 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22333 | HTTPS FTP |
-Validation report
| Summary document |  emd_22333_validation.pdf.gz emd_22333_validation.pdf.gz | 78.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22333_full_validation.pdf.gz emd_22333_full_validation.pdf.gz | 77.2 KB | Display | |
| Data in XML |  emd_22333_validation.xml.gz emd_22333_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22333 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22333 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22333 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22333 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22333.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22333.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SPN3US mottled capsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.512 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
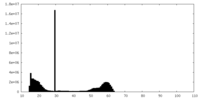
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22333_msk_1.map emd_22333_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
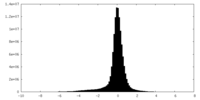
| Density Histograms |
-Additional map: Local resolution map calculated with blocres
| File | emd_22333_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution map calculated with blocres | ||||||||||||
| Projections & Slices |
| ||||||||||||
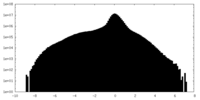
| Density Histograms |
-Half map: half-volume 1
| File | emd_22333_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-volume 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half-volume 2
| File | emd_22333_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-volume 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Salmonella phage SPN3US
| Entire | Name:  Salmonella phage SPN3US (virus) Salmonella phage SPN3US (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Salmonella phage SPN3US
| Supramolecule | Name: Salmonella phage SPN3US / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 1090134 / Sci species name: Salmonella phage SPN3US / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Salmonella enterica (bacteria) Salmonella enterica (bacteria) |
| Host system | Organism:  Salmonella enterica (bacteria) / Recombinant strain: Typhimurium T9079 Salmonella enterica (bacteria) / Recombinant strain: Typhimurium T9079 |
| Molecular weight | Theoretical: 364 MDa |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 1460.0 Å / T number (triangulation number): 27 |
| Virus shell | Shell ID: 2 / Name: Inner shell / Diameter: 1300.0 Å / T number (triangulation number): 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY Details: Grids were from Ted Pella; Lacey carbon with a thin, continuous carbon film |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
| Details | Mottled capsid isolated from a cell lysate and purified using sucrose density gradient centrifugation. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-60 / Number grids imaged: 1 / Number real images: 2244 / Average exposure time: 15.0 sec. / Average electron dose: 138.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 3.1 µm / Calibrated defocus min: 0.57 µm / Calibrated magnification: 28400 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)