[English] 日本語
 Yorodumi
Yorodumi- EMDB-22291: Structure of the human MICU1-MICU2 heterodimer, calcium bound, in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22291 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human MICU1-MICU2 heterodimer, calcium bound, in association with a lipid nanodisc | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / calcium channel / mitochondrial calcium uniporter / MCU / EMRE / mitochondria / CALCIUM BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial crista junction / negative regulation of mitochondrial calcium ion concentration / regulation of cellular hyperosmotic salinity response / positive regulation of cristae formation / mitochondrial calcium ion transmembrane transport / Processing of SMDT1 / uniplex complex / positive regulation of mitochondrial calcium ion concentration / Mitochondrial calcium ion transport / mitochondrial calcium ion homeostasis ...mitochondrial crista junction / negative regulation of mitochondrial calcium ion concentration / regulation of cellular hyperosmotic salinity response / positive regulation of cristae formation / mitochondrial calcium ion transmembrane transport / Processing of SMDT1 / uniplex complex / positive regulation of mitochondrial calcium ion concentration / Mitochondrial calcium ion transport / mitochondrial calcium ion homeostasis / calcium import into the mitochondrion / calcium ion sensor activity / cellular response to calcium ion starvation / calcium ion import / calcium channel inhibitor activity / Mitochondrial protein degradation / calcium channel complex / calcium channel regulator activity / cellular response to calcium ion / defense response / mitochondrial membrane / protein homooligomerization / mitochondrial intermembrane space / mitochondrial inner membrane / protein heterodimerization activity / calcium ion binding / mitochondrion / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Long SB / Wang C | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Structures reveal gatekeeping of the mitochondrial Ca uniporter by MICU1-MICU2. Authors: Chongyuan Wang / Agata Jacewicz / Bryce D Delgado / Rozbeh Baradaran / Stephen Barstow Long /  Abstract: The mitochondrial calcium uniporter is a Ca-gated ion channel complex that controls mitochondrial Ca entry and regulates cell metabolism. MCU and EMRE form the channel while Ca-dependent regulation ...The mitochondrial calcium uniporter is a Ca-gated ion channel complex that controls mitochondrial Ca entry and regulates cell metabolism. MCU and EMRE form the channel while Ca-dependent regulation is conferred by MICU1 and MICU2 through an enigmatic process. We present a cryo-EM structure of an MCU-EMRE-MICU1-MICU2 holocomplex comprising MCU and EMRE subunits from the beetle Tribolium castaneum in complex with a human MICU1-MICU2 heterodimer at 3.3 Å resolution. With analogy to how neuronal channels are blocked by protein toxins, a uniporter interaction domain on MICU1 binds to a channel receptor site comprising MCU and EMRE subunits to inhibit ion flow under resting Ca conditions. A Ca-bound structure of MICU1-MICU2 at 3.1 Å resolution indicates how Ca-dependent changes enable dynamic response to cytosolic Ca signals. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22291.map.gz emd_22291.map.gz | 727.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22291-v30.xml emd-22291-v30.xml emd-22291.xml emd-22291.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22291.png emd_22291.png | 126.5 KB | ||
| Filedesc metadata |  emd-22291.cif.gz emd-22291.cif.gz | 6.3 KB | ||
| Others |  emd_22291_additional.map.gz emd_22291_additional.map.gz | 50.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22291 http://ftp.pdbj.org/pub/emdb/structures/EMD-22291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22291 | HTTPS FTP |
-Validation report
| Summary document |  emd_22291_validation.pdf.gz emd_22291_validation.pdf.gz | 308.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22291_full_validation.pdf.gz emd_22291_full_validation.pdf.gz | 307.7 KB | Display | |
| Data in XML |  emd_22291_validation.xml.gz emd_22291_validation.xml.gz | 6.4 KB | Display | |
| Data in CIF |  emd_22291_validation.cif.gz emd_22291_validation.cif.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22291 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22291 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22291 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22291 | HTTPS FTP |
-Related structure data
| Related structure data |  6xqoMC  6xqnC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22291.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22291.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
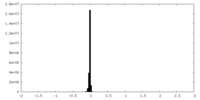
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unsharpened map
| File | emd_22291_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
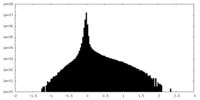
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MICU1-MICU2 complex with calcium
| Entire | Name: MICU1-MICU2 complex with calcium |
|---|---|
| Components |
|
-Supramolecule #1: MICU1-MICU2 complex with calcium
| Supramolecule | Name: MICU1-MICU2 complex with calcium / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Calcium uptake protein 1, mitochondrial
| Macromolecule | Name: Calcium uptake protein 1, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.422883 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPTAAALEPH PEEKKKKRSG FRDRKVMEYE NRIRAYSTPD KIFRYFATLK VISEPGEAEV FMTPEDFVRS ITPNEKQPEH LGLDQYIIK RFDGKKISQE REKFADEGSI FYTLGECGLI SFSDYIFLTT VLSTPQRNFE IAFKMFDLNG DGEVDMEEFE Q VQSIIRSQ ...String: GPTAAALEPH PEEKKKKRSG FRDRKVMEYE NRIRAYSTPD KIFRYFATLK VISEPGEAEV FMTPEDFVRS ITPNEKQPEH LGLDQYIIK RFDGKKISQE REKFADEGSI FYTLGECGLI SFSDYIFLTT VLSTPQRNFE IAFKMFDLNG DGEVDMEEFE Q VQSIIRSQ TSMGMRHRDR PTTGNTLKSG LCSALTTYFF GADLKGKLTI KNFLEFQRKL QHDVLKLEFE RHDPVDGRIT ER QFGGMLL AYSGVQSKKL TAMQRQLKKH FKEGKGLTFQ EVENFFTFLK NINDVDTALS FYHMAGASLD KVTMQQVART VAK VELSDH VCDVVFALFD CDGNGELSNK EFVSIMKQRL MRGLEKPKDM GFTRLMQAMW KCAQETAWDF ALPKQSNW UniProtKB: Calcium uptake protein 1, mitochondrial |
-Macromolecule #2: Calcium uptake protein 2, mitochondrial
| Macromolecule | Name: Calcium uptake protein 2, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.980609 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: HHSRVSVAAR DGSFTVSAQK NVEHGIIYIG KPSLRKQRFM QFSSLEHEGE YYMTPRDFLF SVMFEQMERK TSVKKLTKKD IEDTLSGIQ TAGCGSTFFR DLGDKGLISY TEYLFLLTIL TKPHSGFHVA FKMLDTDGNE MIEKREFFKL QKIISKQDDL M TVKTNETG ...String: HHSRVSVAAR DGSFTVSAQK NVEHGIIYIG KPSLRKQRFM QFSSLEHEGE YYMTPRDFLF SVMFEQMERK TSVKKLTKKD IEDTLSGIQ TAGCGSTFFR DLGDKGLISY TEYLFLLTIL TKPHSGFHVA FKMLDTDGNE MIEKREFFKL QKIISKQDDL M TVKTNETG YQEAIVKEPE INTTLQMRFF GKRGQRKLHY KEFRRFMENL QTEIQEMEFL QFSKGLSFMR KEDFAEWLLF FT NTENKDI YWKNVREKLS AGESISLDEF KSFCHFTTHL EDFAIAMQMF SLAHRPVRLA EFKRAVKVAT GQELSNNILD TVF KIFDLD GDECLSHEEF LGVLKNRMHR GLWVPQHQSI QEYWKCVKKE SIKGVKEVWK QAGKGLF UniProtKB: Calcium uptake protein 2, mitochondrial |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: 2 second blot, blot force of 0. | ||||||||||||
| Details | Monodisperse sample |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Number grids imaged: 5 / Number real images: 21115 / Average exposure time: 4.0 sec. / Average electron dose: 71.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: -3.0 µm / Nominal defocus min: -1.0 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 124 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-6xqo: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)