+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-21654 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Focused asymmetric reconstruction of the hexameric icosahedral 2-fold region of a Heptatis B virus T=4 capsid in complex with the antiviral DBT1 | |||||||||
 マップデータ マップデータ | The sample is a Hepatitis B virus capsid in complex with and antiviral molecule. This map is a focused asymmetric reconstruction of a hexamer of subunits, which surround an icosahedral 2-fold axis. | |||||||||
 試料 試料 |
| |||||||||
| 生物種 |  Hepatitis B virus genotype D subtype adw (ウイルス) Hepatitis B virus genotype D subtype adw (ウイルス) | |||||||||
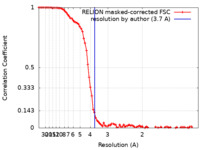
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.7 Å | |||||||||
 データ登録者 データ登録者 | Schlicksup C / Wang JC / Zlotnick A | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: ACS Chem Biol / 年: 2020 ジャーナル: ACS Chem Biol / 年: 2020タイトル: Local Stabilization of Subunit-Subunit Contacts Causes Global Destabilization of Hepatitis B Virus Capsids. 著者: Christopher John Schlicksup / Patrick Laughlin / Steven Dunkelbarger / Joseph Che-Yen Wang / Adam Zlotnick /  要旨: Development of antiviral molecules that bind virion is a strategy that remains in its infancy, and the details of their mechanisms are poorly understood. Here we investigate the behavior of DBT1, a ...Development of antiviral molecules that bind virion is a strategy that remains in its infancy, and the details of their mechanisms are poorly understood. Here we investigate the behavior of DBT1, a dibenzothiazepine that specifically interacts with the capsid protein of hepatitis B virus (HBV). We found that DBT1 stabilizes protein-protein interaction, accelerates capsid assembly, and can induce formation of aberrant particles. Paradoxically, DBT1 can cause preformed capsids to dissociate. These activities may lead to (i) assembly of empty and defective capsids, inhibiting formation of new virus, and (ii) disruption of mature viruses, which are metastable, to inhibit new infection. Using cryo-electron microscopy, we observed that DBT1 led to asymmetric capsids where well-defined DBT1 density was bound at all intersubunit contacts. These results suggest that DBT1 can support assembly by increasing buried surface area but induce disassembly of metastable capsids by favoring asymmetry to induce structural defects. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_21654.map.gz emd_21654.map.gz | 15.8 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-21654-v30.xml emd-21654-v30.xml emd-21654.xml emd-21654.xml | 12.2 KB 12.2 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_21654_fsc.xml emd_21654_fsc.xml | 8.6 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_21654.png emd_21654.png | 67.1 KB | ||
| マスクデータ |  emd_21654_msk_1.map emd_21654_msk_1.map | 52.7 MB |  マスクマップ マスクマップ | |
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21654 http://ftp.pdbj.org/pub/emdb/structures/EMD-21654 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21654 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21654 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_21654_validation.pdf.gz emd_21654_validation.pdf.gz | 78.1 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_21654_full_validation.pdf.gz emd_21654_full_validation.pdf.gz | 77.2 KB | 表示 | |
| XML形式データ |  emd_21654_validation.xml.gz emd_21654_validation.xml.gz | 494 B | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21654 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21654 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21654 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21654 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_21654.map.gz / 形式: CCP4 / 大きさ: 52.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_21654.map.gz / 形式: CCP4 / 大きさ: 52.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | The sample is a Hepatitis B virus capsid in complex with and antiviral molecule. This map is a focused asymmetric reconstruction of a hexamer of subunits, which surround an icosahedral 2-fold axis. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-マスク #1
| ファイル |  emd_21654_msk_1.map emd_21654_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Hepatitis B virus genotype D subtype adw
| 全体 | 名称:  Hepatitis B virus genotype D subtype adw (ウイルス) Hepatitis B virus genotype D subtype adw (ウイルス) |
|---|---|
| 要素 |
|
-超分子 #1: Hepatitis B virus genotype D subtype adw
| 超分子 | 名称: Hepatitis B virus genotype D subtype adw / タイプ: virus / ID: 1 / 親要素: 0 / 含まれる分子: #1 / NCBI-ID: 10419 / 生物種: Hepatitis B virus genotype D subtype adw / Sci species strain: isolate United Kingdom/adyw/1979 / ウイルスタイプ: VIRUS-LIKE PARTICLE / ウイルス・単離状態: OTHER / ウイルス・エンベロープ: No / ウイルス・中空状態: Yes |
|---|---|
| Host system | 生物種:  |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 10 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.5 構成要素:
| |||||||||
| グリッド | 詳細: unspecified | |||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 295.15 K / 装置: FEI VITROBOT MARK III |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: SUPER-RESOLUTION / 撮影したグリッド数: 1 / 実像数: 679 / 平均電子線量: 33.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 初期モデル | PDB ID: Chain - Chain ID: A / Chain - Residue range: 1-143 |
|---|---|
| 精密化 | 空間: REAL / プロトコル: FLEXIBLE FIT / 当てはまり具合の基準: Correlation Coefficient |
 ムービー
ムービー コントローラー
コントローラー













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)