[English] 日本語
 Yorodumi
Yorodumi- EMDB-20954: Cryo-EM structure of human TRPC6 in complex with antagonist AM-1473 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20954 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human TRPC6 in complex with antagonist AM-1473 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRP channel / Antagonist / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of ion transmembrane transporter activity / Role of second messengers in netrin-1 signaling / slit diaphragm / store-operated calcium channel activity / Effects of PIP2 hydrolysis / Elevation of cytosolic Ca2+ levels / cation channel complex / inositol 1,4,5 trisphosphate binding / positive regulation of calcium ion transport / TRP channels ...positive regulation of ion transmembrane transporter activity / Role of second messengers in netrin-1 signaling / slit diaphragm / store-operated calcium channel activity / Effects of PIP2 hydrolysis / Elevation of cytosolic Ca2+ levels / cation channel complex / inositol 1,4,5 trisphosphate binding / positive regulation of calcium ion transport / TRP channels / monoatomic cation transport / single fertilization / regulation of cytosolic calcium ion concentration / monoatomic cation channel activity / calcium channel activity / calcium ion transmembrane transport / positive regulation of cytosolic calcium ion concentration / protein homodimerization activity / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
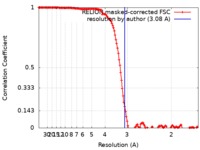
| Method | single particle reconstruction / cryo EM / Resolution: 3.08 Å | |||||||||
 Authors Authors | Bai Y / Yu X | |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Structural basis for pharmacological modulation of the TRPC6 channel. Authors: Yonghong Bai / Xinchao Yu / Hao Chen / Daniel Horne / Ryan White / Xiaosu Wu / Paul Lee / Yan Gu / Sudipa Ghimire-Rijal / Daniel C-H Lin / Xin Huang /  Abstract: Transient receptor potential canonical (TRPC) proteins form nonselective cation channels that play physiological roles in a wide variety of cells. Despite growing evidence supporting the therapeutic ...Transient receptor potential canonical (TRPC) proteins form nonselective cation channels that play physiological roles in a wide variety of cells. Despite growing evidence supporting the therapeutic potential of TRPC6 inhibition in treating pathological cardiac and renal conditions, mechanistic understanding of TRPC6 function and modulation remains obscure. Here we report cryo-EM structures of TRPC6 in both antagonist-bound and agonist-bound states. The structures reveal two novel recognition sites for the small-molecule modulators corroborated by mutagenesis data. The antagonist binds to a cytoplasm-facing pocket formed by S1-S4 and the TRP helix, whereas the agonist wedges at the subunit interface between S6 and the pore helix. Conformational changes upon ligand binding illuminate a mechanistic rationale for understanding TRPC6 modulation. Furthermore, structural and mutagenesis analyses suggest several disease-related mutations enhance channel activity by disrupting interfacial interactions. Our results provide principles of drug action that may facilitate future design of small molecules to ameliorate TRPC6-mediated diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20954.map.gz emd_20954.map.gz | 223.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20954-v30.xml emd-20954-v30.xml emd-20954.xml emd-20954.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20954_fsc.xml emd_20954_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_20954.png emd_20954.png | 183.2 KB | ||
| Filedesc metadata |  emd-20954.cif.gz emd-20954.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20954 http://ftp.pdbj.org/pub/emdb/structures/EMD-20954 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20954 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20954 | HTTPS FTP |
-Validation report
| Summary document |  emd_20954_validation.pdf.gz emd_20954_validation.pdf.gz | 560.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20954_full_validation.pdf.gz emd_20954_full_validation.pdf.gz | 559.9 KB | Display | |
| Data in XML |  emd_20954_validation.xml.gz emd_20954_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_20954_validation.cif.gz emd_20954_validation.cif.gz | 18.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20954 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20954 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20954 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20954 | HTTPS FTP |
-Related structure data
| Related structure data |  6uzaMC  6uz8C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20954.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20954.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TRPC6
| Entire | Name: TRPC6 |
|---|---|
| Components |
|
-Supramolecule #1: TRPC6
| Supramolecule | Name: TRPC6 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Short transient receptor potential channel 6
| Macromolecule | Name: Short transient receptor potential channel 6 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 97.202867 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AYMFSDRSTS LSIEEERFLD AAEYGNIPVV RKMLEECHSL NVNCVDYMGQ NALQLAVANE HLEITELLLK KENLSRVGDA LLLAISKGY VRIVEAILSH PAFAEGKRLA TSPSQSELQQ DDFYAYDEDG TRFSHDVTPI ILAAHCQEYE IVHTLLRKGA R IERPHDYF ...String: AYMFSDRSTS LSIEEERFLD AAEYGNIPVV RKMLEECHSL NVNCVDYMGQ NALQLAVANE HLEITELLLK KENLSRVGDA LLLAISKGY VRIVEAILSH PAFAEGKRLA TSPSQSELQQ DDFYAYDEDG TRFSHDVTPI ILAAHCQEYE IVHTLLRKGA R IERPHDYF CKCNDCNQKQ KHDSFSHSRS RINAYKGLAS PAYLSLSSED PVMTALELSN ELAVLANIEK EFKNDYKKLS MQ CKDFVVG LLDLCRNTEE VEAILNGDVE TLQSGDHGRP NLSRLKLAIK YEVKKFVAHP NCQQQLLSIW YENLSGLRQQ TMA VKFLVV LAVAIGLPFL ALIYWFAPCS KMGKIMRGPF MKFVAHAASF TIFLGLLVMN AADRFEGTKL LPNETSTDNA KQLF RMKTS CFSWMEMLII SWVIGMIWAE CKEIWTQGPK EYLFELWNML DFGMLAIFAA SFIARFMAFW HASKAQSIID ANDTL KDLT KVTLGDNVKY YNLARIKWDP SDPQIISEGL YAIAVVLSFS RIAYILPANE SFGPLQISLG RTVKDIFKFM VIFIMV FVA FMIGMFNLYS YYIGAKQNEA FTTVEESFKT LFWAIFGLSE VKSVVINYNH KFIENIGYVL YGVYNVTMVI VLLNMLI AM INSSFQEIED DADVEWKFAR AKLWFSYFEE GRTLPVPFNL VPSPKSLFYL LLKLKKWISE LFQGHKKGFQ EDAEMNKI N EEKKLGILGS HEDLSKLSLD KKQVGHNKQP SIRSSEDFHL NSFNNPPRQY QKIMKRLIKR YVLQAQIDKE SDEVNEGEL KEIKQDISSL RYELLEEKSQ NTEDLAELIR ELGEKLSMEP NQEETNR UniProtKB: Short transient receptor potential channel 6 |
-Macromolecule #2: 4-({(1R,2R)-2-[(3R)-3-aminopiperidin-1-yl]-2,3-dihydro-1H-inden-1...
| Macromolecule | Name: 4-({(1R,2R)-2-[(3R)-3-aminopiperidin-1-yl]-2,3-dihydro-1H-inden-1-yl}oxy)benzonitrile type: ligand / ID: 2 / Number of copies: 4 / Formula: R0G |
|---|---|
| Molecular weight | Theoretical: 333.427 Da |
| Chemical component information | 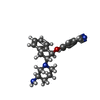 ChemComp-R0G: |
-Macromolecule #3: 2-[[(2~{S})-2-decanoyloxypropoxy]-oxidanyl-phosphoryl]oxyethyl-tr...
| Macromolecule | Name: 2-[[(2~{S})-2-decanoyloxypropoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium type: ligand / ID: 3 / Number of copies: 4 / Formula: S9Y |
|---|---|
| Molecular weight | Theoretical: 396.479 Da |
| Chemical component information | 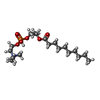 ChemComp-S9Y: |
-Macromolecule #4: [(2~{S})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-octanoyloxy-...
| Macromolecule | Name: [(2~{S})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-octanoyloxy-propan-2-yl] octadecanoate type: ligand / ID: 4 / Number of copies: 4 / Formula: SBJ |
|---|---|
| Molecular weight | Theoretical: 607.8 Da |
| Chemical component information | 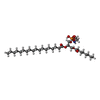 ChemComp-SBJ: |
-Macromolecule #5: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 5 / Number of copies: 8 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)