+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20820 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | A complete structure of the ESX-3 translocon complex | ||||||||||||||||||
 Map data Map data | Consensus map used to generate all focused refinements. Unsharpened | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | ESX / secretion system / type VII secretion system / mycobacteria / complex / membrane protein / TRANSPORT PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on acid anhydrides / hydrolase activity / ATP hydrolysis activity / DNA binding / ATP binding / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||||||||
 Authors Authors | Poweleit N / Rosenberg OS | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: The structure of the endogenous ESX-3 secretion system. Authors: Nicole Poweleit / Nadine Czudnochowski / Rachel Nakagawa / Donovan D Trinidad / Kenan C Murphy / Christopher M Sassetti / Oren S Rosenberg /  Abstract: The ESX (or Type VII) secretion systems are protein export systems in mycobacteria and many Gram-positive bacteria that mediate a broad range of functions including virulence, conjugation, and ...The ESX (or Type VII) secretion systems are protein export systems in mycobacteria and many Gram-positive bacteria that mediate a broad range of functions including virulence, conjugation, and metabolic regulation. These systems translocate folded dimers of WXG100-superfamily protein substrates across the cytoplasmic membrane. We report the cryo-electron microscopy structure of an ESX-3 system, purified using an epitope tag inserted with recombineering into the chromosome of the model organism . The structure reveals a stacked architecture that extends above and below the inner membrane of the bacterium. The ESX-3 protomer complex is assembled from a single copy of the EccB, EccC, and EccE and two copies of the EccD protein. In the structure, the protomers form a stable dimer that is consistent with assembly into a larger oligomer. The ESX-3 structure provides a framework for further study of these important bacterial transporters. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
-Validation report
| Summary document |  emd_20820_validation.pdf.gz emd_20820_validation.pdf.gz | 165.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20820_full_validation.pdf.gz emd_20820_full_validation.pdf.gz | 164.7 KB | Display | |
| Data in XML |  emd_20820_validation.xml.gz emd_20820_validation.xml.gz | 497 B | Display | |
| Data in CIF |  emd_20820_validation.cif.gz emd_20820_validation.cif.gz | 373 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20820 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20820 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20820 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20820 | HTTPS FTP |
-Related structure data
| Related structure data |  6ummMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20820.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20820.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map used to generate all focused refinements. Unsharpened | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
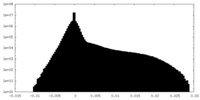
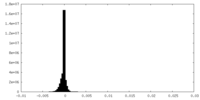
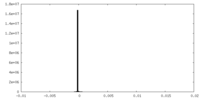
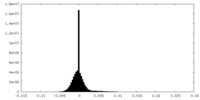
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Mask #1
+Mask #2
+Mask #3
+Mask #4
+Mask #5
+Additional map: Focused refinement of the EccC ATPase 1, 2,...
+Additional map: Half1 of right protomer focused refinement.
+Additional map: Half2 of left protomer focused refinement
+Additional map: Half1 of left protomer focused refinement
+Additional map: Focused refinement of right protomer. Unsharpened
+Additional map: Focused refinement of symmetry expanded protomer. Unsharpened
+Additional map: Half2 of periplasmic focused refinement
+Additional map: Half1 of periplasmic focused refinement
+Additional map: Focused refinement of left protomer. Unsharpened
+Additional map: Focused refinement of periplasmic domain. Unsharpened
+Additional map: Half2 of symmetry expanded protomer
+Additional map: Half1 of symmetry expanded protomer
+Additional map: Half2 of right protomer focused refinement
+Half map: Half2 of consensus map
+Half map: Half1 of consensus map
- Sample components
Sample components
-Entire : ESX-3 translocon complex
| Entire | Name: ESX-3 translocon complex |
|---|---|
| Components |
|
-Supramolecule #1: ESX-3 translocon complex
| Supramolecule | Name: ESX-3 translocon complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 660 KDa |
-Macromolecule #1: ESX-3 secretion system protein EccE3
| Macromolecule | Name: ESX-3 secretion system protein EccE3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 30.813355 KDa |
| Sequence | String: MTARIALASL FVVAAVLAQP WQTTTQRWVL GVSIAAVIVL LAWWKGMFLT TRIGRALAMV RRNRAEDTVE TDAHRATVVL RVDPAAPAQ LPVVVGYLDR YGITCDKVRI THRDAGGTRR SWISLTVDAV DNLAALQARS ARIPLQDTTE VVGRRLADHL R EQGWTVTV ...String: MTARIALASL FVVAAVLAQP WQTTTQRWVL GVSIAAVIVL LAWWKGMFLT TRIGRALAMV RRNRAEDTVE TDAHRATVVL RVDPAAPAQ LPVVVGYLDR YGITCDKVRI THRDAGGTRR SWISLTVDAV DNLAALQARS ARIPLQDTTE VVGRRLADHL R EQGWTVTV VEGVDTPLPV SGKETWRGVA DDAGVVAAYR VKVDDRLDEV LAEIGHLPAE ETWTALEFTG SPAEPLLTVC AA VRTSDRP AAKAPLAGLT PARGRHRPAL AALNPLSTER LDGTAVPL UniProtKB: ESX-3 secretion system protein EccE3 |
-Macromolecule #2: ESX-3 secretion system protein EccD3
| Macromolecule | Name: ESX-3 secretion system protein EccD3 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 48.29248 KDa |
| Sequence | String: MSENTVMPIV RVAVLAAGDD GGRLTEMALP SELPLREILP AVQRIVQPAR ENDGAADPAA APNPVRLSLA PIGGAPFSLD ATLDTVGVV DGDLLALQAV PSGPPAPRIV EDIADAAVIF SEARRRQWGP THIARGAALA LIGLILVGTG LSVAHRVITG D LLGQFIVS ...String: MSENTVMPIV RVAVLAAGDD GGRLTEMALP SELPLREILP AVQRIVQPAR ENDGAADPAA APNPVRLSLA PIGGAPFSLD ATLDTVGVV DGDLLALQAV PSGPPAPRIV EDIADAAVIF SEARRRQWGP THIARGAALA LIGLILVGTG LSVAHRVITG D LLGQFIVS GIALATVIAA LAVRNRSAVL ATSLAVTALV PVAAAFALGV PGDFGAPNVL LAAAGVAAWS LISMAGSPDD RG IAVFTAT AVTGVGVLLV AGAASLWVIS SDVIGCALVL LGLIVTVQAA QLSAMWARFP LPVIPAPGDP TPAARPLSVL ADL PRRVRV SQAHQTGVIA AGVLLGVAGS VALVSSANAS PWAWYIVVAA AAGAALRARV WDSAACKAWL LGHSYLLAVA LLVA FVIGD RYQAALWALA ALAVLVLVWI VAALNPKIAS PDTYSLPMRR MVGFLATGLD ASLIPVMALL VGLFSLVLDR UniProtKB: ESX-3 secretion system protein EccD3 |
-Macromolecule #3: ESX-3 secretion system ATPase EccB3
| Macromolecule | Name: ESX-3 secretion system ATPase EccB3 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number: Hydrolases; Acting on acid anhydrides |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 9.145638 KDa |
| Sequence | String: FSSRTPVNEN PDGVQYRRGF VTRHQVSGWR FVMRRIASGV ALHDTRMLVD PLRTQSRAVL TGALILVTGL VGCFIFSLFR P UniProtKB: ESX-3 secretion system ATPase EccB3 |
-Macromolecule #4: ESX-3 secretion system protein EccC3
| Macromolecule | Name: ESX-3 secretion system protein EccC3 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 44.898371 KDa |
| Sequence | String: MSRLIFEHQR RLTPPTTRKG TITIEPPPQL PRVVPPSLLR RVLPFLIVIL IVGMIVALFA TGMRLISPTM LFFPFVLLLA ATALYRGGD NKMRTEEVDA ERADYLRYLS VVRDNVRAHA AEQRAALEWS HPEPEVLATI PGTRRQWERD PRDRDFLVLR A GRHDVPLD ...String: MSRLIFEHQR RLTPPTTRKG TITIEPPPQL PRVVPPSLLR RVLPFLIVIL IVGMIVALFA TGMRLISPTM LFFPFVLLLA ATALYRGGD NKMRTEEVDA ERADYLRYLS VVRDNVRAHA AEQRAALEWS HPEPEVLATI PGTRRQWERD PRDRDFLVLR A GRHDVPLD AALKVKDTAD EIDLEPVAHS ALRGLLDVQR TVRDAPTGLD VAKLARITVI GEADEARAAI RAWIAQAVTW HD PTMLGVA LAAPDLESGD WSWLKWLPHV DVPNEADGVG PARYLTTSTA ELRERLAPAL ADRPLFPAES GAALKHLLVV LDD PDADPD DIARKPGLTG VTVIHRTTEL PNREQYPDPE RPILRVADGR IERWQVGGWQ PCVDVADAMS AAEAAHIARR LSRW DSN UniProtKB: ESX-3 secretion system protein EccC3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.52 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: Solutions were made fresh and filter-sterilized before use. | ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 7337 / Average electron dose: 73.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6umm: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)