[English] 日本語
 Yorodumi
Yorodumi- EMDB-20651: Structure of the Human Metapneumovirus Polymerase bound to the ph... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20651 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Human Metapneumovirus Polymerase bound to the phosphoprotein tetramer | ||||||||||||
 Map data Map data | None | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | human metapneumovirus / HMPV / Polymerase / Phosphoprotein / Respiratory syncytial virus / RSV / pneumoviridae / Mononegavirales / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationNNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / virion component / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / host cell cytoplasm / RNA-directed RNA polymerase / RNA-dependent RNA polymerase activity / GTPase activity / ATP binding / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Human metapneumovirus CAN97-83 / Human metapneumovirus CAN97-83 /  Human metapneumovirus (strain CAN97-83) Human metapneumovirus (strain CAN97-83) | ||||||||||||
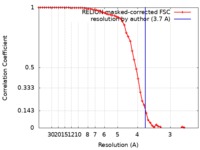
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||
 Authors Authors | Pan J / Qian X | ||||||||||||
| Funding support |  Singapore, Singapore,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structure of the human metapneumovirus polymerase phosphoprotein complex. Authors: Junhua Pan / Xinlei Qian / Simon Lattmann / Abbas El Sahili / Tiong Han Yeo / Huan Jia / Tessa Cressey / Barbara Ludeke / Sarah Noton / Marian Kalocsay / Rachel Fearns / Julien Lescar /   Abstract: Respiratory syncytial virus (RSV) and human metapneumovirus (HMPV) cause severe respiratory diseases in infants and elderly adults. No vaccine or effective antiviral therapy currently exists to ...Respiratory syncytial virus (RSV) and human metapneumovirus (HMPV) cause severe respiratory diseases in infants and elderly adults. No vaccine or effective antiviral therapy currently exists to control RSV or HMPV infections. During viral genome replication and transcription, the tetrameric phosphoprotein P serves as a crucial adaptor between the ribonucleoprotein template and the L protein, which has RNA-dependent RNA polymerase (RdRp), GDP polyribonucleotidyltransferase and cap-specific methyltransferase activities. How P interacts with L and mediates the association with the free form of N and with the ribonucleoprotein is not clear for HMPV or other major human pathogens, including the viruses that cause measles, Ebola and rabies. Here we report a cryo-electron microscopy reconstruction that shows the ring-shaped structure of the polymerase and capping domains of HMPV-L bound to a tetramer of P. The connector and methyltransferase domains of L are mobile with respect to the core. The putative priming loop that is important for the initiation of RNA synthesis is fully retracted, which leaves space in the active-site cavity for RNA elongation. P interacts extensively with the N-terminal region of L, burying more than 4,016 Å of the molecular surface area in the interface. Two of the four helices that form the coiled-coil tetramerization domain of P, and long C-terminal extensions projecting from these two helices, wrap around the L protein in a manner similar to tentacles. The structural versatility of the four P protomers-which are largely disordered in their free state-demonstrates an example of a 'folding-upon-partner-binding' mechanism for carrying out P adaptor functions. The structure shows that P has the potential to modulate multiple functions of L and these results should accelerate the design of specific antiviral drugs. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20651.map.gz emd_20651.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20651-v30.xml emd-20651-v30.xml emd-20651.xml emd-20651.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20651_fsc.xml emd_20651_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_20651.png emd_20651.png | 110 KB | ||
| Filedesc metadata |  emd-20651.cif.gz emd-20651.cif.gz | 8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20651 http://ftp.pdbj.org/pub/emdb/structures/EMD-20651 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20651 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20651 | HTTPS FTP |
-Validation report
| Summary document |  emd_20651_validation.pdf.gz emd_20651_validation.pdf.gz | 577.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20651_full_validation.pdf.gz emd_20651_full_validation.pdf.gz | 577.1 KB | Display | |
| Data in XML |  emd_20651_validation.xml.gz emd_20651_validation.xml.gz | 8.3 KB | Display | |
| Data in CIF |  emd_20651_validation.cif.gz emd_20651_validation.cif.gz | 10.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20651 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20651 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20651 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20651 | HTTPS FTP |
-Related structure data
| Related structure data |  6u5oMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20651.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20651.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
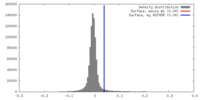
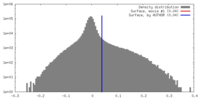
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human metapneumovirus CAN97-83
| Entire | Name:  Human metapneumovirus CAN97-83 Human metapneumovirus CAN97-83 |
|---|---|
| Components |
|
-Supramolecule #1: Human metapneumovirus CAN97-83
| Supramolecule | Name: Human metapneumovirus CAN97-83 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Human metapneumovirus L and P genes were co-expressed in sf9 insect cells using recombinant baculovirus expression system. The protein complex were affinity purified using nickel-NTA resin, ...Details: Human metapneumovirus L and P genes were co-expressed in sf9 insect cells using recombinant baculovirus expression system. The protein complex were affinity purified using nickel-NTA resin, followed by purification on a HiTrap Heparin column and a superpose 6 size exclusion column. |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus CAN97-83 Human metapneumovirus CAN97-83 |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 Details: amino acid residues 1-25 is the N-terminal expression/purification tag containing a Strep II tag, a linker and a TEV protease cleavage site. Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus (strain CAN97-83) / Strain: CAN97-83 Human metapneumovirus (strain CAN97-83) / Strain: CAN97-83 |
| Molecular weight | Theoretical: 233.889562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGWSHPQFEK SSGVDLGTEN LYFQSMDPLN ESTVNVYLPD SYLKGVISFS ETNAIGSCLL KRPYLKNDNT AKVAIENPVI EHVRLKNAV NSKMKISDYK VVEPVNMQHE IMKNVHSCEL TLLKQFLTRS KNISTLKLNM ICDWLQLKST SDDTSILSFI D VEFIPSWV ...String: MGWSHPQFEK SSGVDLGTEN LYFQSMDPLN ESTVNVYLPD SYLKGVISFS ETNAIGSCLL KRPYLKNDNT AKVAIENPVI EHVRLKNAV NSKMKISDYK VVEPVNMQHE IMKNVHSCEL TLLKQFLTRS KNISTLKLNM ICDWLQLKST SDDTSILSFI D VEFIPSWV SNWFSNWYNL NKLILEFRRE EVIRTGSILC RSLGKLVFIV SSYGCIVKSN KSKRVSFFTY NQLLTWKDVM LS RFNANFC IWVSNSLNEN QEGLGLRSNL QGMLTNKLYE TVDYMLSLCC NEGFSLVKEF EGFIMSEILR ITEHAQFSTR FRN TLLNGL TDQLTKLKNK NRLRVHSTVL ENNDYPMYEV VLKLLGDTLR CIKLLINKNL ENAAELYYIF RIFGHPMVDE RDAM DAVKL NNEITKILRL ESLTELRGAF ILRIIKGFVD NNKRWPKIKN LKVLSKRWTM YFKAKNYPSQ LELSEQDFLE LAAIQ FEQE FSVPEKTNLE MVLNDKAISP PKRLIWSVYP KNYLPETIKN RYLEETFNAS DSLKTRRVLE YYLKDNKFDQ KELKSY VVR QEYLNDKEHI VSLTGKEREL SVGRMFAMQP GKQRQIQILA EKLLADNIVP FFPETLTKYG DLDLQRIMEI KSELSSI KT RRNDSYNNYI ARASIVTDLS KFNQAFRYET TAICADVADE LHGTQSLFCW LHLIVPMTTM ICAYRHAPPE TKGEYDID K IEEQSGLYRY HMGGIEGWCQ KLWTMEAISL LDVVSVKTRC QMTSLLNGDN QSIDVSKPVK LSEGLDEVKA DYRLAVKML KEIRDAYRNI GHKLKEGETY ISRDLQFISK VIQSEGVMHP TPIKKVLRVG PWINTILDDI KTSAESIGSL CQELEFRGES IIVSLILRN FWLYNLYMHE SKQHPLAGKQ LFKQLNKTLT SVQRFFEIKR ENEVVDLWMN IPMQFGGGDP VVFYRSFYRR T PDFLTEAI SHVDILLKIS ANIKNETKVS FFKALLSIEK NERATLTTLM RDPQAVGSER QAKVTSDINR TAVTSILSLS PN QLFSDSA IHYSRNEEEV GIIAENITPV YPHGLRVLYE SLPFHKAEKV VNMISGTKSI TNLLQRTSAI NGEDIDRAVS MML ENLGLL SRILSVVVDS IEIPIKSNGR LICCQISRTL RETSWNNMEI VGVTSPSITT CMDVIYATSS HLKGIIIEKF STDR TTRGQ RGPKSPWVGS STQEKKLVPV YNRQILSKQQ REQLEAIGKM RWVYKGTPGL RRLLNKICLG SLGISYKCVK PLLPR FMSV NFLHRLSVSS RPMEFPASVP AYRTTNYHFD TSPINQALSE RFGNEDINLV FQNAISCGIS IMSVVEQLTG RSPKQL VLI PQLEEIDIMP PPVFQGKFNY KLVDKITSDQ HIFSPDKIDM LTLGKMLMPT IKGQKTDQFL NKRENYFHGN NLIESLS AA LACHWCGILT EQCIENNIFK KDWGDGFISD HAFMDFKIFL CVFKTKLLCS WGSQGKNIKD EDIVDESIDK LLRIDNTF W RMFSKVMFEP KVKKRIMLYD VKFLSLVGYI GFKNWFIEQL RSAELHEIPW IVNAEGDLVE IKSIKIYLQL IEQSLFLRI TVLNYTDMAH ALTRLIRKKL MCDNALLTPI SSPMVNLTQV IDPTTQLDYF PKITFERLKN YDTSSNYAKG KLTRNYMILL PWQHVNRYN FVFSSTGCKV SLKTCIGKLM KDLNPKVLYF IGEGAGNWMA RTACEYPDIK FVYRSLKDDL DHHYPLEYQR V IGELSRII DSGEGLSMET TDATQKTHWD LIHRVSKDAL LITLCDAEFK DRDDFFKMVI LWRKHVLSCR ICTTYGTDLY LF AKYHAKD CNVKLPFFVR SVATFIMQGS KLSGSECYIL LTLGHHNSLP CHGEIQNSKM KIAVCNDFYA AKKLDNKSIE ANC KSLLSG LRIPINKKEL DRQRRLLTLQ SNHSSVATVG GSKIIESKWL TNKASTIIDW LEHILNSPKG ELNYDFFEAL ENTY PNMIK LIDNLGNAEI KKLIKVTGYM LVSKK UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: Phosphoprotein
| Macromolecule | Name: Phosphoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus (strain CAN97-83) / Strain: CAN97-83 Human metapneumovirus (strain CAN97-83) / Strain: CAN97-83 |
| Molecular weight | Theoretical: 35.685066 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHHH SSGVDLGTEN LYFQSMSFPE GKDILFMGNE AAKLAEAFQK SLRKPSHKRS QSIIGEKVNT VSETLELPTI SRPTKPTIL SEPKLAWTDK GGAIKTEAKQ TIKVMDPIEE EEFTEKRVLP SSDGKTPAEK KLKPSTNTKK KVSFTPNEPG K YTKLEKDA ...String: MGHHHHHHHH SSGVDLGTEN LYFQSMSFPE GKDILFMGNE AAKLAEAFQK SLRKPSHKRS QSIIGEKVNT VSETLELPTI SRPTKPTIL SEPKLAWTDK GGAIKTEAKQ TIKVMDPIEE EEFTEKRVLP SSDGKTPAEK KLKPSTNTKK KVSFTPNEPG K YTKLEKDA LDLLSDNEEE DAESSILTFE ERDTSSLSIE ARLESIEEKL SMILGLLRTL NIATAGPTAA RDGIRDAMIG IR EELIADI IKEAKGKAAE MMEEEMNQRT KIGNGSVKLT EKAKELNKIV EDESTSGESE EEEELKDTQE NNQEDDIYQL IM UniProtKB: Phosphoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: sample of protein complex was in buffer containing 20 mM Na Hepes, pH 7.4, 300 mM NaCl, 1 mM MgCl2 and 0.5 mM TCEP (components as listed). | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Details: 20 mA | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: 3.5 uL sample (0.8 mg/mL)/grid, 4-second blotting (double-sided) at offset -2 before plunging.. | |||||||||||||||
| Details | purified human metapneumovirus L:P complex in a 1:4 stoichiometry. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 77.0 K / Max: 90.0 K |
| Details | preliminary grid screening was performed manually on a FEI Tecnai F20 microscope equipped with a Gatan K2 summit camera. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7676 pixel / Digitization - Dimensions - Height: 7420 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 7438 / Average exposure time: 0.2 sec. / Average electron dose: 1.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.5 µm / Calibrated magnification: 40323 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 31000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)