+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-20191 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of vaccine-elicited antibody 0PV-b.01 in complex with HIV-1 Env BG505 DS-SOSIP and antibodies VRC03 and PGT122 | |||||||||
 マップデータ マップデータ | Sharpened map | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Fusion Peptide / FP / HIV-1 / SOSIP / Vaccine / IMMUNE SYSTEM | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane 類似検索 - 分子機能 | |||||||||
| 生物種 |   Human immunodeficiency virus 1 (ヒト免疫不全ウイルス) / Human immunodeficiency virus 1 (ヒト免疫不全ウイルス) /  Homo sapiens (ヒト) / Homo sapiens (ヒト) /  | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.5 Å | |||||||||
 データ登録者 データ登録者 | Gorman J / Kwong PD | |||||||||
 引用 引用 |  ジャーナル: Cell / 年: 2019 ジャーナル: Cell / 年: 2019タイトル: Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization. 著者: Rui Kong / Hongying Duan / Zizhang Sheng / Kai Xu / Priyamvada Acharya / Xuejun Chen / Cheng Cheng / Adam S Dingens / Jason Gorman / Mallika Sastry / Chen-Hsiang Shen / Baoshan Zhang / ...著者: Rui Kong / Hongying Duan / Zizhang Sheng / Kai Xu / Priyamvada Acharya / Xuejun Chen / Cheng Cheng / Adam S Dingens / Jason Gorman / Mallika Sastry / Chen-Hsiang Shen / Baoshan Zhang / Tongqing Zhou / Gwo-Yu Chuang / Cara W Chao / Ying Gu / Alexander J Jafari / Mark K Louder / Sijy O'Dell / Ariana P Rowshan / Elise G Viox / Yiran Wang / Chang W Choi / Martin M Corcoran / Angela R Corrigan / Venkata P Dandey / Edward T Eng / Hui Geng / Kathryn E Foulds / Yicheng Guo / Young D Kwon / Bob Lin / Kevin Liu / Rosemarie D Mason / Martha C Nason / Tiffany Y Ohr / Li Ou / Reda Rawi / Edward K Sarfo / Arne Schön / John P Todd / Shuishu Wang / Hui Wei / Winston Wu / / James C Mullikin / Robert T Bailer / Nicole A Doria-Rose / Gunilla B Karlsson Hedestam / Diana G Scorpio / Julie Overbaugh / Jesse D Bloom / Bridget Carragher / Clinton S Potter / Lawrence Shapiro / Peter D Kwong / John R Mascola /   要旨: The vaccine-mediated elicitation of antibodies (Abs) capable of neutralizing diverse HIV-1 strains has been a long-standing goal. To understand how broadly neutralizing antibodies (bNAbs) can be ...The vaccine-mediated elicitation of antibodies (Abs) capable of neutralizing diverse HIV-1 strains has been a long-standing goal. To understand how broadly neutralizing antibodies (bNAbs) can be elicited, we identified, characterized, and tracked five neutralizing Ab lineages targeting the HIV-1-fusion peptide (FP) in vaccinated macaques over time. Genetic and structural analyses revealed two of these lineages to belong to a reproducible class capable of neutralizing up to 59% of 208 diverse viral strains. B cell analysis indicated each of the five lineages to have been initiated and expanded by FP-carrier priming, with envelope (Env)-trimer boosts inducing cross-reactive neutralization. These Abs had binding-energy hotspots focused on FP, whereas several FP-directed Abs induced by immunization with Env trimer-only were less FP-focused and less broadly neutralizing. Priming with a conserved subregion, such as FP, can thus induce Abs with binding-energy hotspots coincident with the target subregion and capable of broad neutralization. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_20191.map.gz emd_20191.map.gz | 157 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-20191-v30.xml emd-20191-v30.xml emd-20191.xml emd-20191.xml | 31.3 KB 31.3 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_20191.png emd_20191.png | 115.3 KB | ||
| マスクデータ |  emd_20191_msk_1.map emd_20191_msk_1.map | 166.4 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-20191.cif.gz emd-20191.cif.gz | 7.9 KB | ||
| その他 |  emd_20191_additional.map.gz emd_20191_additional.map.gz emd_20191_half_map_1.map.gz emd_20191_half_map_1.map.gz emd_20191_half_map_2.map.gz emd_20191_half_map_2.map.gz | 82.5 MB 154.4 MB 154.5 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20191 http://ftp.pdbj.org/pub/emdb/structures/EMD-20191 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20191 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20191 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_20191_validation.pdf.gz emd_20191_validation.pdf.gz | 969 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_20191_full_validation.pdf.gz emd_20191_full_validation.pdf.gz | 968.6 KB | 表示 | |
| XML形式データ |  emd_20191_validation.xml.gz emd_20191_validation.xml.gz | 15 KB | 表示 | |
| CIF形式データ |  emd_20191_validation.cif.gz emd_20191_validation.cif.gz | 17.7 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20191 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20191 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20191 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20191 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  6ot1MC  8977C  9189C  9319C  9320C  9359C  6mpgC  6mphC  6mqcC  6mqeC  6mqmC  6mqrC  6mqsC  6n16C  6n1vC  6n1wC  6nf2C  6osyC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_20191.map.gz / 形式: CCP4 / 大きさ: 166.4 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_20191.map.gz / 形式: CCP4 / 大きさ: 166.4 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.07325 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-マスク #1
| ファイル |  emd_20191_msk_1.map emd_20191_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
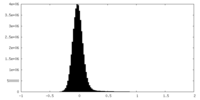
| 密度ヒストグラム |
-追加マップ: Unsharpened map
| ファイル | emd_20191_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Unsharpened map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
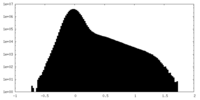
| 密度ヒストグラム |
-ハーフマップ: Half map A
| ファイル | emd_20191_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half map A | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Half map B
| ファイル | emd_20191_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half map B | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : HIV-1 Env BG505 DS-SOSIP and antibodies 0PV-b.01, VRC03 and PGT122
+超分子 #1: HIV-1 Env BG505 DS-SOSIP and antibodies 0PV-b.01, VRC03 and PGT122
+超分子 #2: Envelope glycoprotein gp120
+超分子 #3: Envelope glycoprotein gp41
+超分子 #4: VRC03
+超分子 #5: PGT122
+超分子 #6: 0PV-b.01
+分子 #1: Envelope glycoprotein gp41
+分子 #2: BG505 gp120
+分子 #3: 0PV-b.01 heavy
+分子 #4: 0PV-b.01 light
+分子 #5: PGT122 heavy
+分子 #6: PGT122 light
+分子 #7: VRC03 heavy
+分子 #8: VRC03 light
+分子 #14: 2-acetamido-2-deoxy-beta-D-glucopyranose
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 1.0 mg/mL |
|---|---|
| 緩衝液 | pH: 7.4 / 構成要素 - 式: PBS |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 90 % / チャンバー内温度: 293 K / 装置: SPOTITON |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / 撮影したグリッド数: 1 / 実像数: 2298 / 平均露光時間: 10.0 sec. / 平均電子線量: 73.46 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 70.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)