[English] 日本語
 Yorodumi
Yorodumi- EMDB-19973: Cryo-EM structure of Staphylococcus aureus bacteriophage phi812 b... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Staphylococcus aureus bacteriophage phi812 baseplate in the post-contraction state - sheath initiator, wedge module, and inner tripod proteins | |||||||||
 Map data Map data | Phage phi812 baseplate in the post-contraction state - complete; postprocessed map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteriophage / phage / contractile / phi812 / baseplate / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationDomain of unknown function DUF4815 / Domain of unknown function (DUF4815) / Protein of unknown function DUF2634 / Protein of unknown function (DUF2634) / Baseplate protein J-like / Baseplate J-like protein / LysM domain superfamily / LysM domain Similarity search - Domain/homology | |||||||||
| Biological species |  Staphylococcus phage 812 (virus) Staphylococcus phage 812 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Binovsky J / Siborova M / Baska R / Pichel-Beleiro A / Skubnik K / Novacek J / van Raaij MJ / Plevka P | |||||||||
| Funding support | European Union,  Czech Republic, 2 items Czech Republic, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cell attachment and tail contraction of S. aureus phage phi812 Authors: Binovsky J / Siborova M / Baska R / Pichel-Beleiro A / Skubnik K / Novacek J / van Raaij MJ / Plevka P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19973.map.gz emd_19973.map.gz | 72.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19973-v30.xml emd-19973-v30.xml emd-19973.xml emd-19973.xml | 25.3 KB 25.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19973_fsc.xml emd_19973_fsc.xml | 25.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_19973.png emd_19973.png | 120.5 KB | ||
| Masks |  emd_19973_msk_1.map emd_19973_msk_1.map | 1.4 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-19973.cif.gz emd-19973.cif.gz | 7.9 KB | ||
| Others |  emd_19973_half_map_1.map.gz emd_19973_half_map_1.map.gz emd_19973_half_map_2.map.gz emd_19973_half_map_2.map.gz | 1.1 GB 1.1 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19973 http://ftp.pdbj.org/pub/emdb/structures/EMD-19973 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19973 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19973 | HTTPS FTP |
-Validation report
| Summary document |  emd_19973_validation.pdf.gz emd_19973_validation.pdf.gz | 934.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19973_full_validation.pdf.gz emd_19973_full_validation.pdf.gz | 934.4 KB | Display | |
| Data in XML |  emd_19973_validation.xml.gz emd_19973_validation.xml.gz | 33.5 KB | Display | |
| Data in CIF |  emd_19973_validation.cif.gz emd_19973_validation.cif.gz | 45.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19973 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19973 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19973 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19973 | HTTPS FTP |
-Related structure data
| Related structure data |  9eukMC  9eufC  9eugC  9euhC  9euiC  9eujC  9eulC  9eumC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19973.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19973.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Phage phi812 baseplate in the post-contraction state - complete; postprocessed map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.057 Å | ||||||||||||||||||||||||||||||||||||
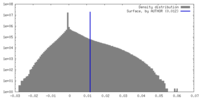
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19973_msk_1.map emd_19973_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19973_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19973_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Staphylococcus phage 812
| Entire | Name:  Staphylococcus phage 812 (virus) Staphylococcus phage 812 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Staphylococcus phage 812
| Supramolecule | Name: Staphylococcus phage 812 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 307898 / Sci species name: Staphylococcus phage 812 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: Yes |
|---|
-Supramolecule #2: Tail
| Supramolecule | Name: Tail / type: complex / ID: 2 / Parent: 1 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage 812 (virus) Staphylococcus phage 812 (virus) |
-Supramolecule #3: Baseplate
| Supramolecule | Name: Baseplate / type: complex / ID: 3 / Parent: 2 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage 812 (virus) Staphylococcus phage 812 (virus) |
-Macromolecule #1: Baseplate wedge subunit
| Macromolecule | Name: Baseplate wedge subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage 812 (virus) Staphylococcus phage 812 (virus) |
| Molecular weight | Theoretical: 26.611752 KDa |
| Sequence | String: MRFKKHVVQH EETMQAIAQR YYGDVSYWID LVEHNNLKYP YLVETDEEKM KDPERLASTG DTLIIPIESD LTDVSAKEIN SRDKDVLVE LALGRDLNIT ADEKYFNEHG TSDNILAFST NGNGDLDTVK GIDNMKQQLQ ARLLTPRGSL MLHPNYGSDL H NLFGLNIP ...String: MRFKKHVVQH EETMQAIAQR YYGDVSYWID LVEHNNLKYP YLVETDEEKM KDPERLASTG DTLIIPIESD LTDVSAKEIN SRDKDVLVE LALGRDLNIT ADEKYFNEHG TSDNILAFST NGNGDLDTVK GIDNMKQQLQ ARLLTPRGSL MLHPNYGSDL H NLFGLNIP EQATLIEMEV LRTLTSDNRV KSANLIDWKI QGNVYSGQFS VEIKSVEESI NFVLGQDEEG IFALFE UniProtKB: Baseplate wedge subunit |
-Macromolecule #2: Baseplate component
| Macromolecule | Name: Baseplate component / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage 812 (virus) Staphylococcus phage 812 (virus) |
| Molecular weight | Theoretical: 39.248859 KDa |
| Sequence | String: MKTRKLTNIL SKLIDKTMAG TSKITDFTPG SASRSLLEAV SLEIEQFYIL TKENIDWGIQ EGIIEAFDFQ KRQSKRAYGD VTIQFYQPL DMRMYIPAGT TFTSTRQEYP QQFETLVDYY AEPDSTEIVV EVYCKETGVA GNVPEGTINT IASGSSLIRS V NNEYSFNT ...String: MKTRKLTNIL SKLIDKTMAG TSKITDFTPG SASRSLLEAV SLEIEQFYIL TKENIDWGIQ EGIIEAFDFQ KRQSKRAYGD VTIQFYQPL DMRMYIPAGT TFTSTRQEYP QQFETLVDYY AEPDSTEIVV EVYCKETGVA GNVPEGTINT IASGSSLIRS V NNEYSFNT GTKEESQEDF KRRFHSFVES RGRATNKSVR YGALQIPDVE GVYVYEETGH ITVFAHDRNG NLSDTLKEDI ID ALQDYRP SGIMLDVTGV EKEEVNVSAT VTISNKSRIG DTLQKHIESV IRSYLNNLKT SDDLIITDLI QAIMNIDDVL IYD VSFDNL DENIIVPPQG IIRAGEIKVE LK UniProtKB: Baseplate component |
-Macromolecule #3: TmpF
| Macromolecule | Name: TmpF / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage 812 (virus) Staphylococcus phage 812 (virus) |
| Molecular weight | Theoretical: 116.391883 KDa |
| Sequence | String: MANFLKNLHP LLRRDRNKKD NQDPNFALID ALNEEMNQVE KDAIESKLQS SLKTSTSEYL DKFGDWFGVY RKTDEKDDVY RARIIKYLL LKRGTNNAII DAIKDYLGRD DIDVSVYEPF TNIFYTNKSH LNGEDHLMGY YYRFAVINVS IGDYFPVEII D VINEFKPA ...String: MANFLKNLHP LLRRDRNKKD NQDPNFALID ALNEEMNQVE KDAIESKLQS SLKTSTSEYL DKFGDWFGVY RKTDEKDDVY RARIIKYLL LKRGTNNAII DAIKDYLGRD DIDVSVYEPF TNIFYTNKSH LNGEDHLMGY YYRFAVINVS IGDYFPVEII D VINEFKPA GVTLYVTYDG ASTIRGGAII KWDDGLPKIE TYQEFDRFTG YDDTFYGHIN MNQSKDTDNS SSDIFKTNHS LI NSLDVLT GSSSVGRQYI NYGYVTSYVY NPGMTSSVNQ ISASTEGRGQ EVPTDYYMYT STKNNNTVEL SMQTTSGVSY LYN NFNFRD YMSKYRPQVD LQSDEARRIV SDYIKELSID YYLSAVIPPD ESIEIKLQVY DFSINRWLTV SINNLSFYEK NIGS NIGYI KDYLNSELNM FTRLEINAGK RDSVDIKVNY LDLMFYYYER GIYTIKPYKA LIENYLDISR ETYVEAFKIA SLSNG DIIT KTGFQPIGYL KLVGNYENTI PSTINIVAKD TDNNPIESNE LDVYNTVENR NLLQSYKGVN TIAREITSTK EFTVSG WAK EIYSTNYLSK VLKPGKVYTL SFDMEITGND PTLKSYSDNH GIYLYSNTKG IVVNGVKSME RTIGNKVSVT QTFTAPT IT DHRLLIYTGR YTSDGKASTP PVFFNTVKIT ELKLTEGSSK LEYSPAPEDK PNVIEKGIKF NNILTNIQTL SINSDTIL K NVTLYYSYYG DSWVELKTLG NISTGETTET NNLIDLYGLQ TVDYSNINPM SKVSLRSIWN VKLGELNNQE GSLSNMPND YFNAVWQDID KLSDIELGSM RMVKDTEGGV FDGATGEIIK ATLFNVGAYT DLDMLAYTLT NYTEPLTLGS SRLISELKEE LLTSESFNV DNRIKVIDSI YEELPNTSII KNGFVEREVT GSKYLDYGLY EPIEDGTRYK LIVEGEFKDN IEFISLYNSN P NFNETFIY PSEIINGVAE KEFIAKPSTE DKPRLNTDVR IYIRPYDSTI SKVRRVELRK V UniProtKB: TmpF |
-Macromolecule #4: DUF4815 domain-containing protein
| Macromolecule | Name: DUF4815 domain-containing protein / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage 812 (virus) Staphylococcus phage 812 (virus) |
| Molecular weight | Theoretical: 129.262961 KDa |
| Sequence | String: MAINFKGSPY LDRFDPSKDR TKVLFNPDRP LQQAELNEMQ SIDQYYLKNL GDAIFKDGDK QSGLGFTLSE DNVLTVNPGY VYINGKIRY YDNDDSVKIT GVGKETIGIK LTERIVTPDE DASLLDQTSG VPSYFSKGAD RLEEKMSLTV NDPTSATIYT F MDGDLYIQ ...String: MAINFKGSPY LDRFDPSKDR TKVLFNPDRP LQQAELNEMQ SIDQYYLKNL GDAIFKDGDK QSGLGFTLSE DNVLTVNPGY VYINGKIRY YDNDDSVKIT GVGKETIGIK LTERIVTPDE DASLLDQTSG VPSYFSKGAD RLEEKMSLTV NDPTSATIYT F MDGDLYIQ STNAEMDKIN KVLAERTYDE SGSYKVNGFE LFSEGNAEDD DHVSVVVDAG KAYVKGFKVD KPVSTRISVP KS YDLGTAE NESTIFNKSN NSISLANSPV KEIRRVTGQV LIEKERVTRG AQGDGQDFLS NNTAFEIVKV WTETSPGVTT KEY KQGEDF RLTDGQTIDW SPQGQEPSGG TSYYVSYKYN KRMEAGKDYE VTTQGEGLSK KWYINFTPSN GAKPIDQTVV LVDY TYYLA RKDSVFINKY GDIAILPGEP NIMRLVTPPL NTDPENLQLG TVTVLPDSDE AVCISFAITR LSMEDLQKVK TRVDN LEYN QAVNALDDGA MEGQNPLTLR SVFSEGFISL DKADITHPDF GIVFSFEDAE ATLAYTEAVN QPKIIPGDTT AHIWGR LIS APFTEERTIY QGQASETLNV NPYNIPNKQG VLKLTPSEDN WIDTENVTIT EQKTKKVTMK RFWRHNESYY GETEHYL YS NLQLDAGQKW KGETYAYDRE HGRTGTLLES GGQRTLEEMI EFIRIRDVSF EVKGLNPNDN NLYLLFDGVR CAITPATG Y RKGSEDGTIM TDAKGTAKGK FTIPAGIRCG NREVTLKNAN STSATTYTAQ GRKKTAQDII IRTRVTVNLV DPLAQSFQY DENRTISSLG LYFASKGDKQ SNVVIQIRGM GDQGYPNKTI YAETVMNADD IKVSNNASAE TRVYFDDPMM AEGGKEYAIV IITENSDYT MWVGTRTKPK IDKPNEVISG NPYLQGVLFS SSNASTWTPH QNSDLKFGIY TSKFNETATI EFEPIKDVSA D RIVLMSTY LTPERTGCTW EMKLILDDMA SSTTFDQLKW EPIGNYQDLD VLGLARQVKL RATFESNRYI SPLMSSSDLT FT TFLTELT GSYVGRAIDM TEAPYNTVRF SYEAFLPKGT KVVPKYSADD GKTWKTFTKS PTTTRANNEF TRYVIDEKVK SSG TNTKLQ VRLDLSTENS FLRPRVRRLM VTTRDE UniProtKB: DUF4815 domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 50mM Tris, 10mM NaCl, 10mM CaCl2 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number real images: 15371 / Average exposure time: 7.0 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-9euk: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)