[English] 日本語
 Yorodumi
Yorodumi- EMDB-19860: Focused map 5 - K48-linked ubiquitin chain formation with a culli... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Focused map 5 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide | |||||||||
 Map data Map data | DeepEMhancer map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CUL2 / FEM1C / ELOBC / SIL1 / Ubiquitin / Ubiquitin Ligase / Ubiquitin chain formation / Poliubiquitylation / LIGASE | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
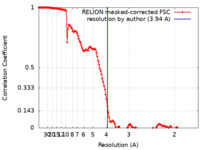
| Method | single particle reconstruction / cryo EM / Resolution: 3.94 Å | |||||||||
 Authors Authors | Liwocha J / Prabu JR / Kleiger G / Schulman BA | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases. Authors: Joanna Liwocha / Jerry Li / Nicholas Purser / Chutima Rattanasopa / Samuel Maiwald / David T Krist / Daniel C Scott / Barbara Steigenberger / J Rajan Prabu / Brenda A Schulman / Gary Kleiger /   Abstract: E3 ubiquitin ligases, in collaboration with E2 ubiquitin-conjugating enzymes, modify proteins with poly-ubiquitin chains. Cullin-RING ligase (CRL) E3s use Cdc34/UBE2R-family E2s to build Lys48-linked ...E3 ubiquitin ligases, in collaboration with E2 ubiquitin-conjugating enzymes, modify proteins with poly-ubiquitin chains. Cullin-RING ligase (CRL) E3s use Cdc34/UBE2R-family E2s to build Lys48-linked poly-ubiquitin chains to control an enormous swath of eukaryotic biology. Yet the molecular mechanisms underlying this exceptional linkage specificity and millisecond kinetics of poly-ubiquitylation remain unclear. Here we obtain cryogenic-electron microscopy (cryo-EM) structures that provide pertinent insight into how such poly-ubiquitin chains are forged. The CRL RING domain not only activates the E2-bound ubiquitin but also shapes the conformation of a distinctive UBE2R2 loop, positioning both the ubiquitin to be transferred and the substrate-linked acceptor ubiquitin within the active site. The structures also reveal how the ubiquitin-like protein NEDD8 uniquely activates CRLs during chain formation. NEDD8 releases the RING domain from the CRL, but unlike previous CRL-E2 structures, does not contact UBE2R2. These findings suggest how poly-ubiquitylation may be accomplished by many E2s and E3s. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19860.map.gz emd_19860.map.gz | 182.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19860-v30.xml emd-19860-v30.xml emd-19860.xml emd-19860.xml | 27.8 KB 27.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19860_fsc.xml emd_19860_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_19860.png emd_19860.png | 50.4 KB | ||
| Masks |  emd_19860_msk_1.map emd_19860_msk_1.map | 209.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19860.cif.gz emd-19860.cif.gz | 6.6 KB | ||
| Others |  emd_19860_additional_1.map.gz emd_19860_additional_1.map.gz emd_19860_half_map_1.map.gz emd_19860_half_map_1.map.gz emd_19860_half_map_2.map.gz emd_19860_half_map_2.map.gz | 9.5 MB 166.4 MB 166.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19860 http://ftp.pdbj.org/pub/emdb/structures/EMD-19860 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19860 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19860 | HTTPS FTP |
-Validation report
| Summary document |  emd_19860_validation.pdf.gz emd_19860_validation.pdf.gz | 813 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19860_full_validation.pdf.gz emd_19860_full_validation.pdf.gz | 812.6 KB | Display | |
| Data in XML |  emd_19860_validation.xml.gz emd_19860_validation.xml.gz | 21.2 KB | Display | |
| Data in CIF |  emd_19860_validation.cif.gz emd_19860_validation.cif.gz | 27.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19860 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19860 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19860 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19860 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19860.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19860.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8512 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19860_msk_1.map emd_19860_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_19860_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19860_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19860_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : K48-linked ubiquitin chain formation with a cullin-RING E3 ligase...
| Entire | Name: K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and CDC34:NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide |
|---|---|
| Components |
|
-Supramolecule #1: K48-linked ubiquitin chain formation with a cullin-RING E3 ligase...
| Supramolecule | Name: K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and CDC34:NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 0.19 kDa/nm |
-Supramolecule #2: RING E3 ligase (RBX1) and Cullin-2 (CUL2)
| Supramolecule | Name: RING E3 ligase (RBX1) and Cullin-2 (CUL2) / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #4, #6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: CDC34, NEDD8, FEM1C with UBE2R2-donor UB-acceptor UB-SIL1 peptide...
| Supramolecule | Name: CDC34, NEDD8, FEM1C with UBE2R2-donor UB-acceptor UB-SIL1 peptide and K48-linked ubiquitin chain type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1-#3, #5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Ubiquitin-conjugating enzyme E2 R2
| Macromolecule | Name: Ubiquitin-conjugating enzyme E2 R2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: E2 ubiquitin-conjugating enzyme |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.190932 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAQQQMTSSQ KALMLELKSL QEEPVEGFRI TLVDESDLYN WEVAIFGPPN TLYEGGYFKA HIKFPIDYPY SPPTFRFLTK MWHPNIYEN GDVCISILHP PVDDPQSGEL PSERWNPTQN VRTILLSVIS LLNEPNTFSP ANVDASVMFR KWRDSKGKDK E YAEIIRKQ ...String: MAQQQMTSSQ KALMLELKSL QEEPVEGFRI TLVDESDLYN WEVAIFGPPN TLYEGGYFKA HIKFPIDYPY SPPTFRFLTK MWHPNIYEN GDVCISILHP PVDDPQSGEL PSERWNPTQN VRTILLSVIS LLNEPNTFSP ANVDASVMFR KWRDSKGKDK E YAEIIRKQ VSATKAEAEK DGVKVPTTLA EYCIKTKVPS NDNSSDLLYD DLYDDDIDDE DEEEEDADCY DDDDSGNEES |
-Macromolecule #2: Polyubiquitin-C,Nucleotide exchange factor SIL1
| Macromolecule | Name: Polyubiquitin-C,Nucleotide exchange factor SIL1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 79.226711 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGCQL EDGRTLSDYN IQKESTLHLV LRLRGGMQIF VKTLTGKTI TLEVEPSDTI ENVKAKIQDK EGIPPDQQRL IFAGKQLEDG RTLSDYNIQK ESTLHLVLRL RGGMQIFVKT L TGKTITLE ...String: MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGCQL EDGRTLSDYN IQKESTLHLV LRLRGGMQIF VKTLTGKTI TLEVEPSDTI ENVKAKIQDK EGIPPDQQRL IFAGKQLEDG RTLSDYNIQK ESTLHLVLRL RGGMQIFVKT L TGKTITLE VEPSDTIENV KAKIQDKEGI PPDQQRLIFA GKQLEDGRTL SDYNIQKEST LHLVLRLRGG MQIFVKTLTG KT ITLEVEP SDTIENVKAK IQDKEGIPPD QQRLIFAGKQ LEDGRTLSDY NIQKESTLHL VLRLRGGMQI FVKTLTGKTI TLE VEPSDT IENVKAKIQD KEGIPPDQQR LIFAGKQLED GRTLSDYNIQ KESTLHLVLR LRGGMQIFVK TLTGKTITLE VEPS DTIEN VKAKIQDKEG IPPDQQRLIF AGKQLEDGRT LSDYNIQKES TLHLVLRLRG GMQIFVKTLT GKTITLEVEP SDTIE NVKA KIQDKEGIPP DQQRLIFAGK QLEDGRTLSD YNIQKESTLH LVLRLRGGMQ IFVKTLTGKT ITLEVEPSDT IENVKA KIQ DKEGIPPDQQ RLIFAGKQLE DGRTLSDYNI QKESTLHLVL RLRGGMQIFV KTLTGKTITL EVEPSDTIEN VKAKIQD KE GIPPDQQRLI FAGKQLEDGR TLSDYNIQKE STLHLVLRLR GGVEGYFQEL LGSVNPTQGR AR |
-Macromolecule #3: Protein fem-1 homolog C
| Macromolecule | Name: Protein fem-1 homolog C / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 68.767312 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDLKTAVFNA ARDGKLRLLT KLLASKSKEE VSSLISEKTN GATPLLMAAR YGHLDMVEFL LEQCSASIEV GGSVNFDGET IEGAPPLWA ASAAGHLKVV QSLLNHGASV NNTTLTNSTP LRAACFDGHL EIVKYLVEHK ADLEVSNRHG HTCLMISCYK G HKEIAQYL ...String: MDLKTAVFNA ARDGKLRLLT KLLASKSKEE VSSLISEKTN GATPLLMAAR YGHLDMVEFL LEQCSASIEV GGSVNFDGET IEGAPPLWA ASAAGHLKVV QSLLNHGASV NNTTLTNSTP LRAACFDGHL EIVKYLVEHK ADLEVSNRHG HTCLMISCYK G HKEIAQYL LEKGADVNRK SVKGNTALHD CAESGSLDIM KMLLMYCAKM EKDGYGMTPL LSASVTGHTN IVDFLTHHAQ TS KTERINA LELLGATFVD KKRDLLGALK YWKKAMNMRY SDRTNIISKP VPQTLIMAYD YAKEVNSAEE LEGLIADPDE MRM QALLIR ERILGPSHPD TSYYIRYRGA VYADSGNFKR CINLWKYALD MQQSNLDPLS PMTASSLLSF AELFSFMLQD RAKG LLGTT VTFDDLMGIL CKSVLEIERA IKQTQCPADP LQLNKALSII LHLICLLEKV PCTLEQDHFK KQTIYRFLKL HPRGK NNFS PLHLAVDKNT TCVGRYPVCK FPSLQVTAIL IECGADVNVR DSDDNSPLHI AALNNHPDIM NLLIKSGAHF DATNLH KQT ASDLLDEKEI AKNLIQPINH TTLQCLAARV IVNHRIYYKG HIPEKLETFV SLHR |
-Macromolecule #4: E3 ubiquitin-protein ligase RBX1
| Macromolecule | Name: E3 ubiquitin-protein ligase RBX1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.289977 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAAAMDVDTP SGTNSGAGKK RFEVKKWNAV ALWAWDIVVD NCAICRNHIM DLCIECQANQ ASATSEECTV AWGVCNHAFH FHCISRWLK TRQVCPLDNR EWEFQKYGH |
-Macromolecule #5: Ubiquitin
| Macromolecule | Name: Ubiquitin / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 77.120398 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGCQL EDGRTLSDYN IQKESTLHLV LRLRGGMQIF VKTLTGKTI TLEVEPSDTI ENVKAKIQDK EGIPPDQQRL IFAGKQLEDG RTLSDYNIQK ESTLHLVLRL RGGMQIFVKT L TGKTITLE ...String: MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGCQL EDGRTLSDYN IQKESTLHLV LRLRGGMQIF VKTLTGKTI TLEVEPSDTI ENVKAKIQDK EGIPPDQQRL IFAGKQLEDG RTLSDYNIQK ESTLHLVLRL RGGMQIFVKT L TGKTITLE VEPSDTIENV KAKIQDKEGI PPDQQRLIFA GKQLEDGRTL SDYNIQKEST LHLVLRLRGG MQIFVKTLTG KT ITLEVEP SDTIENVKAK IQDKEGIPPD QQRLIFAGKQ LEDGRTLSDY NIQKESTLHL VLRLRGGMQI FVKTLTGKTI TLE VEPSDT IENVKAKIQD KEGIPPDQQR LIFAGKQLED GRTLSDYNIQ KESTLHLVLR LRGGMQIFVK TLTGKTITLE VEPS DTIEN VKAKIQDKEG IPPDQQRLIF AGKQLEDGRT LSDYNIQKES TLHLVLRLRG GMQIFVKTLT GKTITLEVEP SDTIE NVKA KIQDKEGIPP DQQRLIFAGK QLEDGRTLSD YNIQKESTLH LVLRLRGGMQ IFVKTLTGKT ITLEVEPSDT IENVKA KIQ DKEGIPPDQQ RLIFAGKQLE DGRTLSDYNI QKESTLHLVL RLRGGMQIFV KTLTGKTITL EVEPSDTIEN VKAKIQD KE GIPPDQQRLI FAGKQLEDGR TLSDYNIQKE STLHLVLRLR GGV |
-Macromolecule #6: Cullin-2
| Macromolecule | Name: Cullin-2 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 87.09893 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSLKPRVVDF DETWNKLLTT IKAVVMLEYV ERATWNDRFS DIYALCVAYP EPLGERLYTE TKIFLENHVR HLHKRVLESE EQVLVMYHR YWEEYSKGAD YMDCLYRYLN TQFIKKNKLT EADLQYGYGG VDMNEPLMEI GELALDMWRK LMVEPLQAIL I RMLLREIK ...String: MSLKPRVVDF DETWNKLLTT IKAVVMLEYV ERATWNDRFS DIYALCVAYP EPLGERLYTE TKIFLENHVR HLHKRVLESE EQVLVMYHR YWEEYSKGAD YMDCLYRYLN TQFIKKNKLT EADLQYGYGG VDMNEPLMEI GELALDMWRK LMVEPLQAIL I RMLLREIK NDRGGEDPNQ KVIHGVINSF VHVEQYKKKF PLKFYQEIFE SPFLTETGEY YKQEASNLLQ ESNCSQYMEK VL GRLKDEE IRCRKYLHPS SYTKVIHECQ QRMVADHLQF LHAECHNIIR QEKKNDMANM YVLLRAVSTG LPHMIQELQN HIH DEGLRA TSNLTQENMP TLFVESVLEV HGKFVQLINT VLNGDQHFMS ALDKALTSVV NYREPKSVCK APELLAKYCD NLLK KSAKG MTENEVEDRL TSFITVFKYI DDKDVFQKFY ARMLAKRLIH GLSMSMDSEE AMINKLKQAC GYEFTSKLHR MYTDM SVSA DLNNKFNNFI KNQDTVIDLG ISFQIYVLQA GAWPLTQAPS STFAIPQELE KSVQMFELFY SQHFSGRKLT WLHYLC TGE VKMNYLGKPY VAMVTTYQMA VLLAFNNSET VSYKELQDST QMNEKELTKT IKSLLDVKMI NHDSEKEDID AESSFSL NM NFSSKRTKFK ITTSMQKDTP QEMEQTRSAV DEDRKMYLQA AIVRIMKARK VLRHNALIQE VISQSRARFN PSISMIKK C IEVLIDKQYI ERSQASADEY SYVA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)