+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
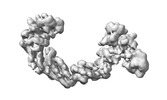
| Title | CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 1 | |||||||||
 Map data Map data | DeepEMhancer sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CUL2 / FEM1C / ELOB/C / SIL1 / Ubiquitin / Ubiquitin Ligase / LIGASE | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
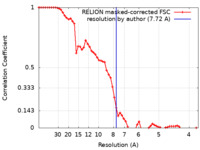
| Method | single particle reconstruction / cryo EM / Resolution: 7.72 Å | |||||||||
 Authors Authors | Liwocha J / Prabu JR / Kleiger G / Schulman BA | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases. Authors: Joanna Liwocha / Jerry Li / Nicholas Purser / Chutima Rattanasopa / Samuel Maiwald / David T Krist / Daniel C Scott / Barbara Steigenberger / J Rajan Prabu / Brenda A Schulman / Gary Kleiger /   Abstract: E3 ubiquitin ligases, in collaboration with E2 ubiquitin-conjugating enzymes, modify proteins with poly-ubiquitin chains. Cullin-RING ligase (CRL) E3s use Cdc34/UBE2R-family E2s to build Lys48-linked ...E3 ubiquitin ligases, in collaboration with E2 ubiquitin-conjugating enzymes, modify proteins with poly-ubiquitin chains. Cullin-RING ligase (CRL) E3s use Cdc34/UBE2R-family E2s to build Lys48-linked poly-ubiquitin chains to control an enormous swath of eukaryotic biology. Yet the molecular mechanisms underlying this exceptional linkage specificity and millisecond kinetics of poly-ubiquitylation remain unclear. Here we obtain cryogenic-electron microscopy (cryo-EM) structures that provide pertinent insight into how such poly-ubiquitin chains are forged. The CRL RING domain not only activates the E2-bound ubiquitin but also shapes the conformation of a distinctive UBE2R2 loop, positioning both the ubiquitin to be transferred and the substrate-linked acceptor ubiquitin within the active site. The structures also reveal how the ubiquitin-like protein NEDD8 uniquely activates CRLs during chain formation. NEDD8 releases the RING domain from the CRL, but unlike previous CRL-E2 structures, does not contact UBE2R2. These findings suggest how poly-ubiquitylation may be accomplished by many E2s and E3s. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17798.map.gz emd_17798.map.gz | 16.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17798-v30.xml emd-17798-v30.xml emd-17798.xml emd-17798.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17798_fsc.xml emd_17798_fsc.xml | 6.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_17798.png emd_17798.png | 41.4 KB | ||
| Masks |  emd_17798_msk_1.map emd_17798_msk_1.map | 18.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17798.cif.gz emd-17798.cif.gz | 4 KB | ||
| Others |  emd_17798_additional_1.map.gz emd_17798_additional_1.map.gz emd_17798_half_map_1.map.gz emd_17798_half_map_1.map.gz emd_17798_half_map_2.map.gz emd_17798_half_map_2.map.gz | 1.3 MB 13.8 MB 13.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17798 http://ftp.pdbj.org/pub/emdb/structures/EMD-17798 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17798 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17798 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17798.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17798.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.885 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17798_msk_1.map emd_17798_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
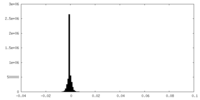
| Density Histograms |
-Additional map: Relion PostProcessed Map
| File | emd_17798_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion PostProcessed Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
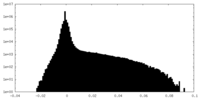
| Density Histograms |
-Half map: #2
| File | emd_17798_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
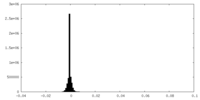
| Density Histograms |
-Half map: #1
| File | emd_17798_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
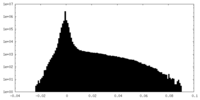
| Density Histograms |
- Sample components
Sample components
-Entire : CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 1
| Entire | Name: CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 1 |
|---|---|
| Components |
|
-Supramolecule #1: CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 1
| Supramolecule | Name: CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 1 / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 190 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.6 µm |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)