[English] 日本語
 Yorodumi
Yorodumi- EMDB-19596: cryo-EM structure of dimerized cross-exon pre-B+5'ss+ATPyS complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of dimerized cross-exon pre-B+5'ss+ATPyS complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | spliceosome / splicing | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 13.0 Å | |||||||||
 Authors Authors | Zhang Z / Kumar V / Stark H / Luehrmann R | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structural insights into the cross-exon to cross-intron spliceosome switch. Authors: Zhenwei Zhang / Vinay Kumar / Olexandr Dybkov / Cindy L Will / Jiayun Zhong / Sebastian E J Ludwig / Henning Urlaub / Berthold Kastner / Holger Stark / Reinhard Lührmann /   Abstract: Early spliceosome assembly can occur through an intron-defined pathway, whereby U1 and U2 small nuclear ribonucleoprotein particles (snRNPs) assemble across the intron. Alternatively, it can occur ...Early spliceosome assembly can occur through an intron-defined pathway, whereby U1 and U2 small nuclear ribonucleoprotein particles (snRNPs) assemble across the intron. Alternatively, it can occur through an exon-defined pathway, whereby U2 binds the branch site located upstream of the defined exon and U1 snRNP interacts with the 5' splice site located directly downstream of it. The U4/U6.U5 tri-snRNP subsequently binds to produce a cross-intron (CI) or cross-exon (CE) pre-B complex, which is then converted to the spliceosomal B complex. Exon definition promotes the splicing of upstream introns and plays a key part in alternative splicing regulation. However, the three-dimensional structure of exon-defined spliceosomal complexes and the molecular mechanism of the conversion from a CE-organized to a CI-organized spliceosome, a pre-requisite for splicing catalysis, remain poorly understood. Here cryo-electron microscopy analyses of human CE pre-B complex and B-like complexes reveal extensive structural similarities with their CI counterparts. The results indicate that the CE and CI spliceosome assembly pathways converge already at the pre-B stage. Add-back experiments using purified CE pre-B complexes, coupled with cryo-electron microscopy, elucidate the order of the extensive remodelling events that accompany the formation of B complexes and B-like complexes. The molecular triggers and roles of B-specific proteins in these rearrangements are also identified. We show that CE pre-B complexes can productively bind in trans to a U1 snRNP-bound 5' splice site. Together, our studies provide new mechanistic insights into the CE to CI switch during spliceosome assembly and its effect on pre-mRNA splice site pairing at this stage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19596.map.gz emd_19596.map.gz | 32.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19596-v30.xml emd-19596-v30.xml emd-19596.xml emd-19596.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19596.png emd_19596.png | 72.5 KB | ||
| Filedesc metadata |  emd-19596.cif.gz emd-19596.cif.gz | 3.7 KB | ||
| Others |  emd_19596_half_map_1.map.gz emd_19596_half_map_1.map.gz emd_19596_half_map_2.map.gz emd_19596_half_map_2.map.gz | 27.2 MB 27.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19596 http://ftp.pdbj.org/pub/emdb/structures/EMD-19596 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19596 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19596 | HTTPS FTP |
-Validation report
| Summary document |  emd_19596_validation.pdf.gz emd_19596_validation.pdf.gz | 749.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19596_full_validation.pdf.gz emd_19596_full_validation.pdf.gz | 748.7 KB | Display | |
| Data in XML |  emd_19596_validation.xml.gz emd_19596_validation.xml.gz | 11.1 KB | Display | |
| Data in CIF |  emd_19596_validation.cif.gz emd_19596_validation.cif.gz | 13.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19596 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19596 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19596 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19596 | HTTPS FTP |
-Related structure data
| Related structure data |  8qozC  8qp8C  8qp9C  8qpaC  8qpbC  8qpeC  8qpkC  8qxdC  8qzsC  8r08C  8r09C  8r0aC  8r0bC  8rm5C C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19596.map.gz / Format: CCP4 / Size: 35.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19596.map.gz / Format: CCP4 / Size: 35.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.48 Å | ||||||||||||||||||||||||||||||||||||
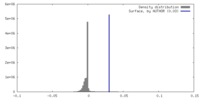
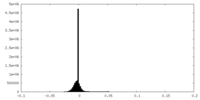
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_19596_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
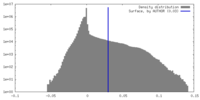
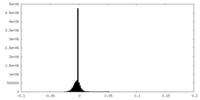
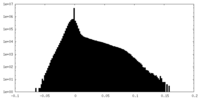
| Density Histograms |
-Half map: #2
| File | emd_19596_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
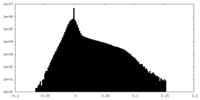
| Density Histograms |
- Sample components
Sample components
-Entire : dimerized pre-B+5'ss+ATPyS particles
| Entire | Name: dimerized pre-B+5'ss+ATPyS particles |
|---|---|
| Components |
|
-Supramolecule #1: dimerized pre-B+5'ss+ATPyS particles
| Supramolecule | Name: dimerized pre-B+5'ss+ATPyS particles / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: ab initio 3D |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 13.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 26552 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


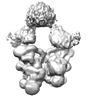


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)