+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1897 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Reconstruction of the 3D model of AMPK trimer in basal state | |||||||||
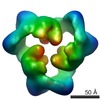
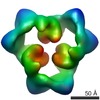
 Map data Map data | This is an image of a surface rendered side-view of AMP-activated protein kinase. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Zhu L / Chen L / Zhou XM / Zhang YY / Zhang YJ / Zhao J / Ji SR / Wu JW / Wu Y | |||||||||
 Citation Citation |  Journal: Structure / Year: 2011 Journal: Structure / Year: 2011Title: Structural insights into the architecture and allostery of full-length AMP-activated protein kinase. Authors: Li Zhu / Lei Chen / Xiao-Ming Zhou / Yuan-Yuan Zhang / Yi-Jiong Zhang / Jing Zhao / Shang-Rong Ji / Jia-Wei Wu / Yi Wu /  Abstract: AMP-activated protein kinase (AMPK) is a heterotrimeric complex composed of α catalytic subunit, β scaffolding subunit, and γ regulatory subunit with critical roles in maintaining cellular energy ...AMP-activated protein kinase (AMPK) is a heterotrimeric complex composed of α catalytic subunit, β scaffolding subunit, and γ regulatory subunit with critical roles in maintaining cellular energy homeostasis. However, the molecular architecture of the intact complex and the allostery associated with the adenosine binding-induced regulation of kinase activity remain unclear. Here, we determine the three-dimensional reconstruction and subunit organization of the full-length rat AMPK (α1β1γ1) through single-particle electron-microscopy. By comparing the structures of AMPK in ATP- and AMP-bound states, we are able to visualize the sequential conformational changes underlying kinase activation that transmits from the adenosine binding sites in the γ subunit to the kinase domain of the α subunit. These results not only make substantial revision to the current model of AMPK assembly, but also highlight a central role of the linker sequence of the α subunit in mediating the allostery of AMPK. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1897.map.gz emd_1897.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1897-v30.xml emd-1897-v30.xml emd-1897.xml emd-1897.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_1897.png emd_1897.png | 139.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1897 http://ftp.pdbj.org/pub/emdb/structures/EMD-1897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1897 | HTTPS FTP |
-Validation report
| Summary document |  emd_1897_validation.pdf.gz emd_1897_validation.pdf.gz | 200.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1897_full_validation.pdf.gz emd_1897_full_validation.pdf.gz | 199.5 KB | Display | |
| Data in XML |  emd_1897_validation.xml.gz emd_1897_validation.xml.gz | 5.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1897 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1897 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1897 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1897 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1897.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1897.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is an image of a surface rendered side-view of AMP-activated protein kinase. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Full-Length Rat AMP-Activated Protein Kinase
| Entire | Name: Full-Length Rat AMP-Activated Protein Kinase |
|---|---|
| Components |
|
-Supramolecule #1000: Full-Length Rat AMP-Activated Protein Kinase
| Supramolecule | Name: Full-Length Rat AMP-Activated Protein Kinase / type: sample / ID: 1000 / Oligomeric state: Homotrimer / Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 390 KDa / Theoretical: 390 KDa |
-Macromolecule #1: Mammalian AMP-Activated Protein Kinase
| Macromolecule | Name: Mammalian AMP-Activated Protein Kinase / type: protein_or_peptide / ID: 1 / Name.synonym: Mammalian AMPK / Number of copies: 3 / Oligomeric state: Homotrimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 390 KDa / Theoretical: 390 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL |
|---|---|
| Buffer | pH: 6.8 / Details: 10mM Tris,15mM NaCl, 1mM TCEP,1.2mM MgCl2 |
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein floated on 1% w/v uranyl acetate for 30 seconds. |
| Grid | Details: 300 mesh copper EM grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 21.17 µm / Number real images: 136 / Average electron dose: 18 e/Å2 / Bits/pixel: 16 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 1.7 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Single tilt / Specimen holder model: OTHER / Tilt angle min: -45 |
- Image processing
Image processing
| Details | The particle pairs were extracted using WEB with a box size of 72 pixels. |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER Details: High-quality classes without significant differences in initial reconstructions were merged for calculating new volumes. Number images used: 3284 |
| Final angle assignment | Details: SPIDER:theta 45 degrees |
| Final two d classification | Number classes: 10 |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)
























