+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Capsid structure of Giardiavirus (GLV) CAT strain | ||||||||||||
 Map data Map data | Full map of GLV-CAT capsid | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Giardia parsite / dsRNA virus / capsid structure / Totiviridae / Giardia lambia virus (GLV) / VIRUS | ||||||||||||
| Function / homology | Capsid protein Function and homology information Function and homology information | ||||||||||||
| Biological species |  Giardia lamblia virus Giardia lamblia virus | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | ||||||||||||
 Authors Authors | Wang H / Gianluca M / Munke A / Hassan MM / Lalle M / Okamoto K | ||||||||||||
| Funding support |  Sweden, Sweden,  Norway, 3 items Norway, 3 items
| ||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2024 Journal: PLoS Pathog / Year: 2024Title: High-resolution comparative atomic structures of two Giardiavirus prototypes infecting G. duodenalis parasite. Authors: Han Wang / Gianluca Marucci / Anna Munke / Mohammad Maruf Hassan / Marco Lalle / Kenta Okamoto /    Abstract: The Giardia lamblia virus (GLV) is a non-enveloped icosahedral dsRNA and endosymbiont virus that infects the zoonotic protozoan parasite Giardia duodenalis (syn. G. lamblia, G. intestinalis), which ...The Giardia lamblia virus (GLV) is a non-enveloped icosahedral dsRNA and endosymbiont virus that infects the zoonotic protozoan parasite Giardia duodenalis (syn. G. lamblia, G. intestinalis), which is a pathogen of mammals, including humans. Elucidating the transmission mechanism of GLV is crucial for gaining an in-depth understanding of the virulence of the virus in G. duodenalis. GLV belongs to the family Totiviridae, which infects yeast and protozoa intracellularly; however, it also transmits extracellularly, similar to the phylogenetically, distantly related toti-like viruses that infect multicellular hosts. The GLV capsid structure is extensively involved in the longstanding discussion concerning extracellular transmission in Totiviridae and toti-like viruses. Hence, this study constructed the first high-resolution comparative atomic models of two GLV strains, namely GLV-HP and GLV-CAT, which showed different intracellular localization and virulence phenotypes, using cryogenic electron microscopy single-particle analysis. The atomic models of the GLV capsids presented swapped C-terminal extensions, extra surface loops, and a lack of cap-snatching pockets, similar to those of toti-like viruses. However, their open pores and absence of the extra crown protein resemble those of other yeast and protozoan Totiviridae viruses, demonstrating the essential structures for extracellular cell-to-cell transmission. The structural comparison between GLV-HP and GLV-CAT indicates the first evidence of critical structural motifs for the transmission and virulence of GLV in G. duodenalis. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18792.map.gz emd_18792.map.gz | 937.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18792-v30.xml emd-18792-v30.xml emd-18792.xml emd-18792.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18792.png emd_18792.png | 266.2 KB | ||
| Filedesc metadata |  emd-18792.cif.gz emd-18792.cif.gz | 6 KB | ||
| Others |  emd_18792_half_map_1.map.gz emd_18792_half_map_1.map.gz emd_18792_half_map_2.map.gz emd_18792_half_map_2.map.gz | 928.8 MB 928.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18792 http://ftp.pdbj.org/pub/emdb/structures/EMD-18792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18792 | HTTPS FTP |
-Validation report
| Summary document |  emd_18792_validation.pdf.gz emd_18792_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18792_full_validation.pdf.gz emd_18792_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_18792_validation.xml.gz emd_18792_validation.xml.gz | 22.2 KB | Display | |
| Data in CIF |  emd_18792_validation.cif.gz emd_18792_validation.cif.gz | 26.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18792 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18792 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18792 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18792 | HTTPS FTP |
-Related structure data
| Related structure data |  8r0gMC  8r0fC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18792.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18792.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map of GLV-CAT capsid | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
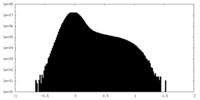
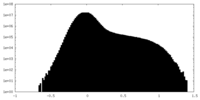
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A of GLV-CAT capsid
| File | emd_18792_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of GLV-CAT capsid | ||||||||||||
| Projections & Slices |
| ||||||||||||
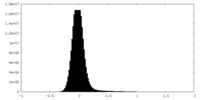
| Density Histograms |
-Half map: Half map B of GLV-CAT capsid
| File | emd_18792_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of GLV-CAT capsid | ||||||||||||
| Projections & Slices |
| ||||||||||||
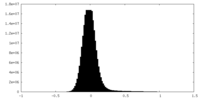
| Density Histograms |
- Sample components
Sample components
-Entire : Giardia lamblia virus
| Entire | Name:  Giardia lamblia virus Giardia lamblia virus |
|---|---|
| Components |
|
-Supramolecule #1: Giardia lamblia virus
| Supramolecule | Name: Giardia lamblia virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: GLV isolated from G. duodenalis isolates CAT-1 from a cat NCBI-ID: 29255 / Sci species name: Giardia lamblia virus / Sci species strain: CAT-1 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 450.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Giardia lamblia virus Giardia lamblia virus |
| Molecular weight | Theoretical: 103.492406 KDa |
| Sequence | String: PENITLDTLN TQNDYEETHG ESPEVPKASI APAGRQNVPV LQQNQENKDN HALGGSEDAK DEREIRFSAI KTLYNYYSEG PSTPIMPHL VNRLRGLDAL AKIDATLTKV DMNAAYIFAL RPTFPYSYGY KQRFSNRRLT TSAVCYARTG LSSFLTVNKT Y TSNSPLKG ...String: PENITLDTLN TQNDYEETHG ESPEVPKASI APAGRQNVPV LQQNQENKDN HALGGSEDAK DEREIRFSAI KTLYNYYSEG PSTPIMPHL VNRLRGLDAL AKIDATLTKV DMNAAYIFAL RPTFPYSYGY KQRFSNRRLT TSAVCYARTG LSSFLTVNKT Y TSNSPLKG GSRGWPIFNV GVSPHVAEPH MRTLSPIGLE VFSQNLATFS KTLLTASSKV FTQSLYTADI LSIFGEVFLP HV MQPVSNY TPILVRALLA LIHILGPGSG NCSLSSSIFE SSIPQFFTVS HSTNMSNRTR YCLHTWSAYK DMFRNGIPPQ STL PPTLAP EGSSARVLIP AALVTSPMFP WLLALVSSGP QFFLYSKDAS INTVDIGSRG RITSPIPDVA NLDLHRLWNL FRFD GHRYI DVVIVGVDRD YVWPYQNGVY VHGGKGPNGT DNYGNADVHD GIGTIFSSFN DNVNVQTSDL ILGLLTLWNH ITTTY ATEE EVTMAIKIAA AFALVYPVQP IVYSGCPRAF QSHTSYYQPS SENCYTTDTA EVKSTWDTVE LSVQVNNAMV LGMTLP FGQ PTVSSAQWFN NIDKAEISMF KVGNLPLQNL DYLSLDMMEF YAPTTGQLYD IRSDNLILSA HRTVNLGIGY TALADFF AY LASVPAQSFY HDRMVTSPIS KQTYSVYERF IERFVDDFVG WDRCDLFNLD TLLAAKRIAG VASSPIPWHC SLQRCPLP I IMHYTGLHFG QEHITVRDVA GVEGLQQIVM RNDQGKIVVD ALGTAAPSRL AVKLDWSRLS AWYSDTTCAI PLSDRVMEI INYAAIWDPT QERRATGFVY TYFSPNFLSS FNASEPIFNT SINLTPPYDD TSHTVIQNLS MPQMLSFDPY YESTFYVVSA DNEWIPTSG PAWKVPYLEN VVKRSGRRLL AELRIASNNG SGDRTFLDDV UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 1 x PBS (-) |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | Purified GLV-CAT particles |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 5804 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)