+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
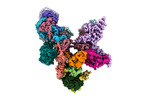
| タイトル | Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba | ||||||||||||||||||
 マップデータ マップデータ | Composite Map (Map G). | ||||||||||||||||||
 試料 試料 |
| ||||||||||||||||||
 キーワード キーワード | Transcription / Chloroplasts / Gene Expression / RNA / Polymerase | ||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報: / DNA-directed RNA polymerase complex / chloroplast / ribonucleoside binding / : / : / : / : / : / : ...: / DNA-directed RNA polymerase complex / chloroplast / ribonucleoside binding / : / : / : / : / : / : / DNA-directed RNA polymerase / protein dimerization activity / magnesium ion binding / mitochondrion / DNA binding / zinc ion binding 類似検索 - 分子機能 | ||||||||||||||||||
| 生物種 |  Sinapis alba (シロガラシ) Sinapis alba (シロガラシ) | ||||||||||||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.5 Å | ||||||||||||||||||
 データ登録者 データ登録者 | do Prado PFV / Ahrens FM / Pfannschmidt T / Hillen HS | ||||||||||||||||||
| 資金援助 |  ドイツ, 5件 ドイツ, 5件
| ||||||||||||||||||
 引用 引用 |  ジャーナル: Mol Cell / 年: 2024 ジャーナル: Mol Cell / 年: 2024タイトル: Structure of the multi-subunit chloroplast RNA polymerase. 著者: Paula F V do Prado / Frederik M Ahrens / Monique Liebers / Noah Ditz / Hans-Peter Braun / Thomas Pfannschmidt / Hauke S Hillen /  要旨: Chloroplasts contain a dedicated genome that encodes subunits of the photosynthesis machinery. Transcription of photosynthesis genes is predominantly carried out by a plastid-encoded RNA polymerase ...Chloroplasts contain a dedicated genome that encodes subunits of the photosynthesis machinery. Transcription of photosynthesis genes is predominantly carried out by a plastid-encoded RNA polymerase (PEP), a nearly 1 MDa complex composed of core subunits with homology to eubacterial RNA polymerases (RNAPs) and at least 12 additional chloroplast-specific PEP-associated proteins (PAPs). However, the architecture of this complex and the functions of the PAPs remain unknown. Here, we report the cryo-EM structure of a 19-subunit PEP complex from Sinapis alba (white mustard). The structure reveals that the PEP core resembles prokaryotic and nuclear RNAPs but contains chloroplast-specific features that mediate interactions with the PAPs. The PAPs are unrelated to known transcription factors and arrange around the core in a unique fashion. Their structures suggest potential functions during transcription in the chemical environment of chloroplasts. These results reveal structural insights into chloroplast transcription and provide a framework for understanding photosynthesis gene expression. | ||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_18496.map.gz emd_18496.map.gz | 291 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-18496-v30.xml emd-18496-v30.xml emd-18496.xml emd-18496.xml | 36.1 KB 36.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_18496.png emd_18496.png | 72.8 KB | ||
| Filedesc metadata |  emd-18496.cif.gz emd-18496.cif.gz | 12.3 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18496 http://ftp.pdbj.org/pub/emdb/structures/EMD-18496 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18496 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18496 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_18496_validation.pdf.gz emd_18496_validation.pdf.gz | 515.5 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_18496_full_validation.pdf.gz emd_18496_full_validation.pdf.gz | 515 KB | 表示 | |
| XML形式データ |  emd_18496_validation.xml.gz emd_18496_validation.xml.gz | 7.3 KB | 表示 | |
| CIF形式データ |  emd_18496_validation.cif.gz emd_18496_validation.cif.gz | 8.5 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18496 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18496 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18496 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18496 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8qmaMC C: 同じ文献を引用 ( M: このマップから作成された原子モデル |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_18496.map.gz / 形式: CCP4 / 大きさ: 325 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_18496.map.gz / 形式: CCP4 / 大きさ: 325 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Composite Map (Map G). | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
+全体 : Plastid-encoded DNA-dependent RNA polymerase (PEP)
+超分子 #1: Plastid-encoded DNA-dependent RNA polymerase (PEP)
+分子 #1: PAP4
+分子 #2: PAP5
+分子 #3: PAP8
+分子 #4: PAP9
+分子 #5: PAP10
+分子 #6: PAP11
+分子 #7: PAP12 (DNA-directed RNA polymerase subunit omega)
+分子 #8: PTAC18
+分子 #9: PAP6
+分子 #10: DNA-directed RNA polymerase subunit beta
+分子 #11: DNA-directed RNA polymerase subunit beta''
+分子 #12: DNA-directed RNA polymerase subunit alpha
+分子 #13: PAP1
+分子 #14: PAP3
+分子 #15: FLN2
+分子 #16: PAP7
+分子 #17: DNA-directed RNA polymerase subunit beta'
+分子 #18: FE (III) ION
+分子 #19: ZINC ION
+分子 #20: S-ADENOSYLMETHIONINE
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.6 構成要素:
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| グリッド | モデル: Quantifoil R2/1 / 材質: COPPER | |||||||||||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 95 % / チャンバー内温度: 277.15 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均露光時間: 2.7 sec. / 平均電子線量: 40.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: OTHER / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 0.5 µm / 倍率(公称値): 81000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| 初期モデル | モデルのタイプ: INSILICO MODEL |
|---|---|
| 最終 再構成 | アルゴリズム: BACK PROJECTION / 解像度のタイプ: BY AUTHOR / 解像度: 3.5 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 使用した粒子像数: 123874 |
| 初期 角度割当 | タイプ: MAXIMUM LIKELIHOOD |
| 最終 角度割当 | タイプ: MAXIMUM LIKELIHOOD |
 ムービー
ムービー コントローラー
コントローラー











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















