[English] 日本語
 Yorodumi
Yorodumi- EMDB-17318: In situ subtomogram average of Prototype Foamy Virus Env hexamer ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ subtomogram average of Prototype Foamy Virus Env hexamer of trimers | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane fusion / envelope glycoprotein / foamy virus / MPER / transmembrane / MEMBRANE PROTEIN / VIRAL PROTEIN | |||||||||
| Biological species |  Eastern chimpanzee simian foamy virus Eastern chimpanzee simian foamy virus | |||||||||
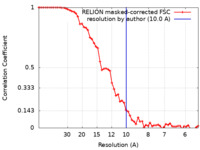
| Method | subtomogram averaging / cryo EM / Resolution: 10.0 Å | |||||||||
 Authors Authors | Calcraft T / Nans A / Rosenthal PB | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry. Authors: Thomas Calcraft / Nicole Stanke-Scheffler / Andrea Nans / Dirk Lindemann / Ian A Taylor / Peter B Rosenthal /   Abstract: Foamy viruses (FVs) are an ancient lineage of retroviruses, with an evolutionary history spanning over 450 million years. Vector systems based on Prototype Foamy Virus (PFV) are promising candidates ...Foamy viruses (FVs) are an ancient lineage of retroviruses, with an evolutionary history spanning over 450 million years. Vector systems based on Prototype Foamy Virus (PFV) are promising candidates for gene and oncolytic therapies. Structural studies of PFV contribute to the understanding of the mechanisms of FV replication, cell entry and infection, and retroviral evolution. Here we combine cryoEM and cryoET to determine high-resolution in situ structures of the PFV icosahedral capsid (CA) and envelope glycoprotein (Env), including its type III transmembrane anchor and membrane-proximal external region (MPER), and show how they are organized in an integrated structure of assembled PFV particles. The atomic models reveal an ancient retroviral capsid architecture and an unexpected relationship between Env and other class 1 fusion proteins of the Mononegavirales. Our results represent the de novo structure determination of an assembled retrovirus particle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17318.map.gz emd_17318.map.gz | 13.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17318-v30.xml emd-17318-v30.xml emd-17318.xml emd-17318.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17318_fsc.xml emd_17318_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_17318.png emd_17318.png | 60.2 KB | ||
| Masks |  emd_17318_msk_1.map emd_17318_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17318.cif.gz emd-17318.cif.gz | 7 KB | ||
| Others |  emd_17318_half_map_1.map.gz emd_17318_half_map_1.map.gz emd_17318_half_map_2.map.gz emd_17318_half_map_2.map.gz | 7.1 MB 7.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17318 http://ftp.pdbj.org/pub/emdb/structures/EMD-17318 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17318 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17318 | HTTPS FTP |
-Validation report
| Summary document |  emd_17318_validation.pdf.gz emd_17318_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17318_full_validation.pdf.gz emd_17318_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_17318_validation.xml.gz emd_17318_validation.xml.gz | 11.5 KB | Display | |
| Data in CIF |  emd_17318_validation.cif.gz emd_17318_validation.cif.gz | 15.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17318 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17318 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17318 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17318 | HTTPS FTP |
-Related structure data
| Related structure data |  8ozqMC  8ozhC  8ozjC  8ozkC  8ozlC  8ozmC  8oznC  8ozpC C: citing same article ( M: atomic model generated by this map |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17318.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17318.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.76 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17318_msk_1.map emd_17318_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17318_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17318_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Eastern chimpanzee simian foamy virus
| Entire | Name:  Eastern chimpanzee simian foamy virus Eastern chimpanzee simian foamy virus |
|---|---|
| Components |
|
-Supramolecule #1: Eastern chimpanzee simian foamy virus
| Supramolecule | Name: Eastern chimpanzee simian foamy virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 2170195 / Sci species name: Eastern chimpanzee simian foamy virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Envelope glycoprotein
| Macromolecule | Name: Envelope glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Eastern chimpanzee simian foamy virus Eastern chimpanzee simian foamy virus |
| Molecular weight | Theoretical: 113.804594 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAPPMTLQQW IIWNKMNKAH EALQNTTTVT EQQKEQIILD IQNEEVQPTR RDKFRYLLYT CCATSSRVLA WIFLVCILLI IVLVSCFVT ISRIQWNKDI QVLGPVIDWN VTQRAVYQPL QTRRIARSLR MQHPVPKYVE VNMTSIPQGV YYEPHPEPIV V KERVLGLS ...String: MAPPMTLQQW IIWNKMNKAH EALQNTTTVT EQQKEQIILD IQNEEVQPTR RDKFRYLLYT CCATSSRVLA WIFLVCILLI IVLVSCFVT ISRIQWNKDI QVLGPVIDWN VTQRAVYQPL QTRRIARSLR MQHPVPKYVE VNMTSIPQGV YYEPHPEPIV V KERVLGLS QILMINSENI ANNANLTQEV KKLLTEMVNE EMQSLSDVMI DFEIPLGDPR DQEQYIHRKC YQEFANCYLV KY KEPKPWP KEGLIADQCP LPGYHAGLTY NRQSIWDYYI KVESIRPANW TTKSKYGQAR LGSFYIPSSL RQINVSHVLF CSD QLYSKW YNIENTIEQN ERFLLNKLNN LTSGTSVLKK RALPKDWSSQ GKNALFREIN VLDICSKPES VILLNTSYYS FSLW EGDCN FTKDMISQLV PECDGFYNNS KWMHMHPYAC RFWRSKNEKE ETKCRDGETK RCLYYPLWDS PESTYDFGYL AYQKN FPSP ICIEQQKIRD QDYEVYSLYQ ECKIASKAYG IDTVLFSLKN FLNYTGTPVN EMPNARAFVG LIDPKFPPSY PNVTRE HYT SCNNRKRRSV DNNYAKLRSM GYALTGAVQT LSQISDINDE NLQQGIYLLR DHVITLMEAT LHDISVMEGM FAVQHLH TH LNHLKTMLLE RRIDWTYMSS TWLQQQLQKS DDEMKVIKRI ARSLVYYVKQ THSSPTATAW EIGLYYELVI PKHIYLNN W NVVNIGHLVK SAGQLTHVTI AHPYEIINKE CVETIYLHLE DCTRQDYVIC DVVKIVQPCG NSSDTSDCPV WAEAVKEPF VQVNPLKNGS YLVLASSTDC QIPPYVPSIV TVNETTSCFG LDFKRPLVAE ERLSFEPRLP NLQLRLPHLV GIIAKIKGIK IEVTSSGES IKEQIERAKA ELLRLDIHEG DTPAWIQQLA AATKDVWPAA ASALQGIGNF LSGTAQGIFG TAFSLLGYLK P ILIGVGVI LLVILIFKIV SWIPTKKKNQ |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #9: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 9 / Number of copies: 3 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #10: PHOSPHATIDYLETHANOLAMINE
| Macromolecule | Name: PHOSPHATIDYLETHANOLAMINE / type: ligand / ID: 10 / Number of copies: 3 / Formula: PTY |
|---|---|
| Molecular weight | Theoretical: 734.039 Da |
| Chemical component information |  ChemComp-PTY: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Average electron dose: 2.62 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 2.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8ozq: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)