[English] 日本語
 Yorodumi
Yorodumi- EMDB-16956: Cryo-EM reconstruction of VP4 assembly from SA11 Rotavirus Non-Tr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of VP4 assembly from SA11 Rotavirus Non-Tripsinized Triple Layered Particle | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Rotavirus / dsRNA virus / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell rough endoplasmic reticulum / permeabilization of host organelle membrane involved in viral entry into host cell / host cytoskeleton / viral outer capsid / host cell endoplasmic reticulum-Golgi intermediate compartment / virion attachment to host cell / host cell plasma membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Rotavirus / Rotavirus /  Rotavirus A Rotavirus A | |||||||||
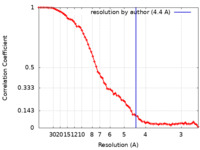
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Asensio-Cob D / Perez-Mata C / Gomez-Blanco J / Vargas J / Rodriguez JM / Luque D | |||||||||
| Funding support |  Spain, 1 items Spain, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural basis of rotavirus spike proteolytic activation Authors: Asensio-Cob D / Perez-Mata C / Gomez-Blanco J / Vargas J / Rodriguez JM / Luque D | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16956.map.gz emd_16956.map.gz | 27.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16956-v30.xml emd-16956-v30.xml emd-16956.xml emd-16956.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16956_fsc.xml emd_16956_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_16956.png emd_16956.png | 66.9 KB | ||
| Filedesc metadata |  emd-16956.cif.gz emd-16956.cif.gz | 6 KB | ||
| Others |  emd_16956_half_map_1.map.gz emd_16956_half_map_1.map.gz emd_16956_half_map_2.map.gz emd_16956_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16956 http://ftp.pdbj.org/pub/emdb/structures/EMD-16956 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16956 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16956 | HTTPS FTP |
-Validation report
| Summary document |  emd_16956_validation.pdf.gz emd_16956_validation.pdf.gz | 639.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16956_full_validation.pdf.gz emd_16956_full_validation.pdf.gz | 638.7 KB | Display | |
| Data in XML |  emd_16956_validation.xml.gz emd_16956_validation.xml.gz | 12.8 KB | Display | |
| Data in CIF |  emd_16956_validation.cif.gz emd_16956_validation.cif.gz | 17.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16956 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16956 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16956 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16956 | HTTPS FTP |
-Related structure data
| Related structure data |  8oleMC  8olbC  8olcC  8qtzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16956.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16956.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||
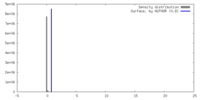
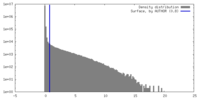
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16956_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
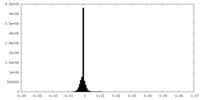
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16956_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
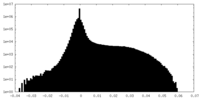
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rotavirus A
| Entire | Name:  Rotavirus A Rotavirus A |
|---|---|
| Components |
|
-Supramolecule #1: Rotavirus A
| Supramolecule | Name: Rotavirus A / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Rotavirus SA11 Non-tripsinized spike / NCBI-ID: 28875 / Sci species name: Rotavirus A / Sci species strain: SA11 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Chlorocebus aethiops (grivet) Chlorocebus aethiops (grivet) |
| Molecular weight | Theoretical: 92 MDa |
| Virus shell | Shell ID: 1 / Name: TLP / Diameter: 700.0 Å |
-Macromolecule #1: Outer capsid protein VP4
| Macromolecule | Name: Outer capsid protein VP4 / type: protein_or_peptide / ID: 1 / Details: Outer capsid protein VP4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rotavirus Rotavirus |
| Molecular weight | Theoretical: 86.749891 KDa |
| Recombinant expression | Organism:  Chlorocebus aethiops (grivet) Chlorocebus aethiops (grivet) |
| Sequence | String: MASLIYRQLL TNSYTVDLSD EIQEIGSTKS QNVTINPGPF AQTGYAPVNW GPGEINDSTT VEPLLDGPYQ PTTFNPPVDY WMLLAPTTP GVIVEGTNNT DRWLATILIE PNVQSENRTY TIFGIQEQLT VSNTSQDQWK FIDVVKTTAN GSIGQYGSLL S SPKLYAVM ...String: MASLIYRQLL TNSYTVDLSD EIQEIGSTKS QNVTINPGPF AQTGYAPVNW GPGEINDSTT VEPLLDGPYQ PTTFNPPVDY WMLLAPTTP GVIVEGTNNT DRWLATILIE PNVQSENRTY TIFGIQEQLT VSNTSQDQWK FIDVVKTTAN GSIGQYGSLL S SPKLYAVM KHNEKLYTYE GQTPNARTGH YSTTNYDSVN MTAFCDFYII PRSEESKCTE YINNGLPPIQ NTRNVVPLSL TA RDVIHYR AQANEDIVIS KTSLWKEMQY NRDITIRFKF ANTIIKSGGL GYKWSEISFK PANYQYTYTR DGEEVTAHTT CSV NGVNDF SFNGGSLPTD FVVSKFEVIK ENSYVYIDYW DDSQAFRNVV YVRSLAANLN SVMCTGGSYN FSLPVGQWPV LTGG AVSLH SAGVTLSTQF TDFVSLNSLR FRFRLAVEEP HFKLTRTRLD RLYGLPAADP NNGKEYYEIA GRFSLISLVP SNDDY QTPI ANSVTVRQDL ERQLGELREE FNALSQEIAM SQLIDLALLP LDMFSMFSGI KSTIDAAKSM ATNVMKKFKK SGLANS VST LTDSLSDAAS SISRGSSIRS IGSSASAWTD VSTQITDISS SVSSVSTQTS TISRRLRLKE MATQTEGMNF DDISAAV LK TKIDKSTQIS PNTIPDIVTE ASEKFIPNRA YRVINNDDVF EAGIDGKFFA YKVDTFEEIP FDVQKFADLV TDSPVISA I IDFKTLKNLN DNYGITKQQA FNLLRSDPRV LREFINQDNP IIRNRIEQLI MQCRL UniProtKB: Outer capsid protein VP4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 295 K / Instrument: LEICA EM GP |
| Details | Rotavirus SA11 Non-tripsinized spike |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Number real images: 1465 / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)