+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SA11 Rotavirus Trypsinized Triple Layered Particle | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Rotavirus / dsRNA virus / virus | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral intermediate capsid / host cell endoplasmic reticulum lumen / T=13 icosahedral viral capsid / T=2 icosahedral viral capsid / viral inner capsid / viral outer capsid / viral nucleocapsid / receptor-mediated virion attachment to host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane ...viral intermediate capsid / host cell endoplasmic reticulum lumen / T=13 icosahedral viral capsid / T=2 icosahedral viral capsid / viral inner capsid / viral outer capsid / viral nucleocapsid / receptor-mediated virion attachment to host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / viral envelope / structural molecule activity / RNA binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Rotavirus / Rotavirus /  Rotavirus A Rotavirus A | |||||||||
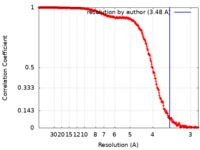
| Method | single particle reconstruction / cryo EM / Resolution: 3.48 Å | |||||||||
 Authors Authors | Asensio-Cob D / Perez-Mata C / Gomez-Blanco J / Vargas J / Rodriguez JM / Luque D | |||||||||
| Funding support |  Spain, 1 items Spain, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2025 Journal: PLoS Pathog / Year: 2025Title: Structural determinants of rotavirus proteolytic activation. Authors: Dunia Asensio-Cob / Carlos P Mata / Josue Gomez-Blanco / Javier Vargas / Javier M Rodriguez / Daniel Luque /    Abstract: The infectivity of rotavirus (RV), the leading cause of childhood diarrhea, hinges on the activation of viral particles through the proteolysis of the spike protein by trypsin-like proteases in the ...The infectivity of rotavirus (RV), the leading cause of childhood diarrhea, hinges on the activation of viral particles through the proteolysis of the spike protein by trypsin-like proteases in the host intestinal lumen. In order to determine the structural basis of trypsin activation, we have used cryogenic electron microscopy (cryo-EM) and advanced image processing methods to compare uncleaved and cleaved RV particles. We find that the conformation of the non-proteolyzed spike is constrained by the position of loops that surround its structure, linking the lectin domains of the spike head to its body. The proteolysis of these loops removes this structural constraint, thereby enabling the spike to undergo the necessary conformational changes required for cell membrane penetration. Thus, these loops function as regulatory elements to ensure that the spike protein is activated precisely when and where it is needed to facilitate a successful infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16955.map.gz emd_16955.map.gz | 1.2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16955-v30.xml emd-16955-v30.xml emd-16955.xml emd-16955.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16955_fsc.xml emd_16955_fsc.xml | 36.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_16955.png emd_16955.png | 216.5 KB | ||
| Filedesc metadata |  emd-16955.cif.gz emd-16955.cif.gz | 7.5 KB | ||
| Others |  emd_16955_half_map_1.map.gz emd_16955_half_map_1.map.gz emd_16955_half_map_2.map.gz emd_16955_half_map_2.map.gz | 1.3 GB 1.3 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16955 http://ftp.pdbj.org/pub/emdb/structures/EMD-16955 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16955 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16955 | HTTPS FTP |
-Related structure data
| Related structure data |  8olcMC  8olbC  8oleC  8qtzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16955.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16955.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.43 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16955_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16955_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rotavirus A
| Entire | Name:  Rotavirus A Rotavirus A |
|---|---|
| Components |
|
-Supramolecule #1: Rotavirus A
| Supramolecule | Name: Rotavirus A / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / Details: Rotavirus SA11 Trypsinized TLP / NCBI-ID: 28875 / Sci species name: Rotavirus A / Sci species strain: SA11 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Chlorocebus aethiops (grivet) Chlorocebus aethiops (grivet) |
| Molecular weight | Theoretical: 92 MDa |
| Virus shell | Shell ID: 1 / Name: TLP / Diameter: 1000.0 Å |
-Macromolecule #1: Outer capsid glycoprotein VP7
| Macromolecule | Name: Outer capsid glycoprotein VP7 / type: protein_or_peptide / ID: 1 / Details: Outer capsid glycoprotein VP7 / Number of copies: 13 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rotavirus Rotavirus |
| Molecular weight | Theoretical: 37.222602 KDa |
| Recombinant expression | Organism:  Chlorocebus aethiops (grivet) Chlorocebus aethiops (grivet) |
| Sequence | String: MYGIEYTTVL TFLISIILLN YILKSLTRIM DFIIYRFLFI IVILSPFLRA QNYGINLPIT GSMDTAYANS TQEETFLTST LCLYYPTEA ATEINDNSWK DTLSQLFLTK GWPTGSVYFK EYTNIASFSV DPQLYCDYNV VLMKYDATLQ LDMSELADLI L NEWLCNPM ...String: MYGIEYTTVL TFLISIILLN YILKSLTRIM DFIIYRFLFI IVILSPFLRA QNYGINLPIT GSMDTAYANS TQEETFLTST LCLYYPTEA ATEINDNSWK DTLSQLFLTK GWPTGSVYFK EYTNIASFSV DPQLYCDYNV VLMKYDATLQ LDMSELADLI L NEWLCNPM DITLYYYQQT DEANKWISMG SSCTIKVCPL NTQTLGIGCL TTDATTFEEV ATAEKLVITD VVDGVNHKLD VT TATCTIR NCKKLGPREN VAVIQVGGSD ILDITADPTT APQTERMMRI NWKKWWQVFY TVVDYVDQII QVMSKRSRSL NSA AFYYRV UniProtKB: Outer capsid glycoprotein VP7 |
-Macromolecule #2: Intermediate capsid protein VP6
| Macromolecule | Name: Intermediate capsid protein VP6 / type: protein_or_peptide / ID: 2 / Details: Intermediate capsid protein VP6 / Number of copies: 13 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rotavirus Rotavirus |
| Molecular weight | Theoretical: 44.910738 KDa |
| Recombinant expression | Organism:  Chlorocebus aethiops (grivet) Chlorocebus aethiops (grivet) |
| Sequence | String: MDVLYSLSKT LKDARDKIVE GTLYSNVSDL IQQFNQMIIT MNGNEFQTGG IGNLPIRNWN FNFGLLGTTL LNLDANYVET ARNTIDYFV DFVDNVCMDE MVRESQRNGI APQSDSLRKL SAIKFKRINF DNSSEYIENW NLQNRRQRTG FTFHKPNIFP Y SASFTLNR ...String: MDVLYSLSKT LKDARDKIVE GTLYSNVSDL IQQFNQMIIT MNGNEFQTGG IGNLPIRNWN FNFGLLGTTL LNLDANYVET ARNTIDYFV DFVDNVCMDE MVRESQRNGI APQSDSLRKL SAIKFKRINF DNSSEYIENW NLQNRRQRTG FTFHKPNIFP Y SASFTLNR SQPAHDNLMG TMWLNAGSEI QVAGFDYSCA INAPANIQQF EHIVPLRRVL TTATITLLPD AERFSFPRVI NS ADGATTW FFNPVILRPN NVEVEFLLNG QIINTYQARF GTIVARNFDT IRLSFQLMRP PNMTPAVAVL FPNAQPFEHH ATV GLTLRI ESAVCESVLA DASETLLANV TSVRQEYAIP VGPVFPPGMN WTDLITNYSP SREDNLQRVF TVASIRSMLI K UniProtKB: Intermediate capsid protein VP6 |
-Macromolecule #3: Inner capsid protein VP2
| Macromolecule | Name: Inner capsid protein VP2 / type: protein_or_peptide / ID: 3 / Details: Inner core protein / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rotavirus Rotavirus |
| Molecular weight | Theoretical: 102.853406 KDa |
| Recombinant expression | Organism:  Chlorocebus aethiops (grivet) Chlorocebus aethiops (grivet) |
| Sequence | String: MAYRKRGARR ETNLKQDERM QEKEDSKNIN NDSPKSQLSE KVLSKKEEII TDNQEEVKIS DEVKKSNKEE SKQLLEVLKT KEEHQKEVQ YEILQKTIPT FEPKESILKK LEDIKPEQAK KQTKLFRIFE PKQLPIYRAN GERELRNRWY WKLKRDTLPD G DYDVREYF ...String: MAYRKRGARR ETNLKQDERM QEKEDSKNIN NDSPKSQLSE KVLSKKEEII TDNQEEVKIS DEVKKSNKEE SKQLLEVLKT KEEHQKEVQ YEILQKTIPT FEPKESILKK LEDIKPEQAK KQTKLFRIFE PKQLPIYRAN GERELRNRWY WKLKRDTLPD G DYDVREYF LNLYDQVLME MPDYLLLKDM AVENKNSRDA GKVVDSETAA ICDAIFQDEE TEGAVRRFIA EMRQRVQADR NV VNYPSIL HPIDHAFNEY FLQHQLVEPL NNDIIFNYIP ERIRNDVNYI LNMDRNLPST ARYIRPNLLQ DRLNLHDNFE SLW DTITTS NYILARSVVP DLKELVSTEA QIQKMSQDLQ LEALTIQSET QFLTGINSQA ANDCFKTLIA AMLSQRTMSL DFVT TNYMS LISGMWLLTV IPNDMFIRES LVACQLAIIN TIVYPAFGMQ RMHYRNGDPQ TPFQIAEQQI QNFQVANWLH FVNYN QFRQ VVIDGVLNQV LNDNIRNGHV VNQLMEALMQ LSRQQFPTMP VDYKRSIQRG ILLLSNRLGQ LVDLTRLLSY NYETLM ACI TMNMQHVQTL TTEKLQLTSV TSLCMLIGNA TVIPSPQTLF HYYNVNVNFH SNYNERINDA VAIITAANRL NLYQKKM KS IVEDFLKRLQ IFDVARVPDD QMYRLRDRLR LLPVEIRRLD IFNLIAMNME QIERASDKIA QGVIIAYRDM QLERDEMY G YVNIARNLDG FQQINLEELM RSGDYAQITN MLLNNQPVAL VGALPFITDS SVISLIAKLD ATVFAQIVKL RKVDTLKPI LYKINSDSND FYLVANYDWI PTSTTKVYKQ VPQQFDFRAS MHMLTSNLTF TVYSDLLAFV SADTVEPINA VAFDNMRIMN EL UniProtKB: Inner capsid protein VP2 |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 26 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 10 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 5 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 295 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number real images: 2368 / Average electron dose: 39.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 58000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)