+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Mitochondrial Lon Y394F Mutant ADP Bound | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | Human mitochondrial AAA+ protease / motor protein / HYDROLASE | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationoxidation-dependent protein catabolic process / PH domain binding / mitochondrial protein catabolic process / G-quadruplex DNA binding / endopeptidase La / mitochondrial DNA metabolic process / mitochondrial genome maintenance / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / mitochondrial nucleoid ...oxidation-dependent protein catabolic process / PH domain binding / mitochondrial protein catabolic process / G-quadruplex DNA binding / endopeptidase La / mitochondrial DNA metabolic process / mitochondrial genome maintenance / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / mitochondrial nucleoid / insulin receptor substrate binding / chaperone-mediated protein complex assembly / DNA polymerase binding / regulation of peptidyl-tyrosine phosphorylation / negative regulation of insulin receptor signaling pathway / Mitochondrial protein degradation / proteolysis involved in protein catabolic process / mitochondrion organization / ADP binding / protein catabolic process / single-stranded DNA binding / cellular response to oxidative stress / sequence-specific DNA binding / single-stranded RNA binding / response to hypoxia / mitochondrial matrix / serine-type endopeptidase activity / ATP hydrolysis activity / mitochondrion / nucleoplasm / ATP binding / identical protein binding / membrane / cytosol Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||
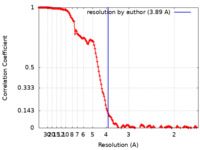
| Method | single particle reconstruction / cryo EM / Resolution: 3.89 Å | ||||||||||||||||||||||||
 Authors Authors | Kereiche S / Bauer JA / Matyas P / Novacek J / Kutejova E | ||||||||||||||||||||||||
| Funding support |  Slovakia, European Union, Slovakia, European Union,  Czech Republic, 7 items Czech Republic, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2024 Journal: Sci Rep / Year: 2024Title: Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions. Authors: Nina Kunová / Gabriela Ondrovičová / Jacob A Bauer / Veronika Krajčovičová / Matyáš Pinkas / Barbora Stojkovičová / Henrieta Havalová / Veronika Lukáčová / Lenka Kohútová / ...Authors: Nina Kunová / Gabriela Ondrovičová / Jacob A Bauer / Veronika Krajčovičová / Matyáš Pinkas / Barbora Stojkovičová / Henrieta Havalová / Veronika Lukáčová / Lenka Kohútová / Július Košťan / Lucia Martináková / Peter Baráth / Jiří Nováček / Sebastian Zoll / Sami Kereϊche / Eva Kutejová / Vladimír Pevala /    Abstract: Phosphorylation plays a crucial role in the regulation of many fundamental cellular processes. Phosphorylation levels are increased in many cancer cells where they may promote changes in ...Phosphorylation plays a crucial role in the regulation of many fundamental cellular processes. Phosphorylation levels are increased in many cancer cells where they may promote changes in mitochondrial homeostasis. Proteomic studies on various types of cancer identified 17 phosphorylation sites within the human ATP-dependent protease Lon, which degrades misfolded, unassembled and oxidatively damaged proteins in mitochondria. Most of these sites were found in Lon's N-terminal (NTD) and ATPase domains, though little is known about the effects on their function. By combining the biochemical and cryo-electron microscopy studies, we show the effect of Tyr186 and Tyr394 phosphorylations in Lon's NTD, which greatly reduce all Lon activities without affecting its ability to bind substrates or perturbing its tertiary structure. A substantial reduction in Lon's activities is also observed in the presence of polyphosphate, whose amount significantly increases in cancer cells. Our study thus provides an insight into the possible fine-tuning of Lon activities in human diseases, which highlights Lon's importance in maintaining proteostasis in mitochondria. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16923.map.gz emd_16923.map.gz | 140.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16923-v30.xml emd-16923-v30.xml emd-16923.xml emd-16923.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16923_fsc.xml emd_16923_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_16923.png emd_16923.png | 144.5 KB | ||
| Filedesc metadata |  emd-16923.cif.gz emd-16923.cif.gz | 6.2 KB | ||
| Others |  emd_16923_half_map_1.map.gz emd_16923_half_map_1.map.gz emd_16923_half_map_2.map.gz emd_16923_half_map_2.map.gz | 261.9 MB 261.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16923 http://ftp.pdbj.org/pub/emdb/structures/EMD-16923 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16923 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16923 | HTTPS FTP |
-Validation report
| Summary document |  emd_16923_validation.pdf.gz emd_16923_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16923_full_validation.pdf.gz emd_16923_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_16923_validation.xml.gz emd_16923_validation.xml.gz | 22.9 KB | Display | |
| Data in CIF |  emd_16923_validation.cif.gz emd_16923_validation.cif.gz | 29.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16923 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16923 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16923 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16923 | HTTPS FTP |
-Related structure data
| Related structure data |  8okaMC  8ojlC  8om7C  8ovfC  8ovgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16923.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16923.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.783 Å | ||||||||||||||||||||||||||||||||||||
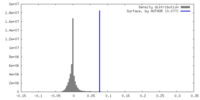
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_16923_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
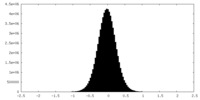
| Density Histograms |
-Half map: #1
| File | emd_16923_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
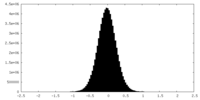
| Density Histograms |
- Sample components
Sample components
-Entire : Human mitochondrial Lon protease
| Entire | Name: Human mitochondrial Lon protease |
|---|---|
| Components |
|
-Supramolecule #1: Human mitochondrial Lon protease
| Supramolecule | Name: Human mitochondrial Lon protease / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organelle: mitochondria Homo sapiens (human) / Organelle: mitochondria |
-Macromolecule #1: Lon protease homolog, mitochondrial
| Macromolecule | Name: Lon protease homolog, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: endopeptidase La |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 98.183852 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHDY DIPTTENLYF QGAHMTIPDV FPHLPLIAIT RNPVFPRFIK IIEVKNKKLV ELLRRKVRLA QPYVGVFLKR DDSNESDVV ESLDEIYHTG TFAQIHEMQD LGDKLRMIVM GHRRVHISRQ LEVEPEEPEA ENKHKPRRKS KRGKKEAEDE L SARHPAEL ...String: MGHHHHHHDY DIPTTENLYF QGAHMTIPDV FPHLPLIAIT RNPVFPRFIK IIEVKNKKLV ELLRRKVRLA QPYVGVFLKR DDSNESDVV ESLDEIYHTG TFAQIHEMQD LGDKLRMIVM GHRRVHISRQ LEVEPEEPEA ENKHKPRRKS KRGKKEAEDE L SARHPAEL AMEPTPELPA EVLMVEVENV VHEDFQVTEE VKALTAEIVK TIRDIIALNP LYRESVLQMM QAGQRVVDNP IY LSDMGAA LTGAESHELQ DVLEETNIPK RLYKALSLLK KEFELSKLQQ RLGREVEEKI KQTHRKFLLQ EQLKIIKKEL GLE KDDKDA IEEKFRERLK ELVVPKHVMD VVDEELSKLG LLDNHSSEFN VTRNYLDWLT SIPWGKYSNE NLDLARAQAV LEED HYGME DVKKRILEFI AVSQLRGSTQ GKILCFYGPP GVGKTSIARS IARALNREYF RFSVGGMTDV AEIKGHRRTY VGAMP GKII QCLKKTKTEN PLILIDEVDK IGRGYQGDPS SALLELLDPE QNANFLDHYL DVPVDLSKVL FICTANVTDT IPEPLR DRM EMINVSGYVA QEKLAIAERY LVPQARALCG LDESKAKLSS DVLTLLIKQY CRESGVRNLQ KQVEKVLRKS AYKIVSG EA ESVEVTPENL QDFVGKPVFT VERMYDVTPP GVVMGLAWTA MGGSTLFVET SLRRPQDKDA KGDKDGSLEV TGQLGEVM K ESARIAYTFA RAFLMQHAPA NDYLVTSHIH LHVPEGATPK DGPSAGCTIV TALLSLAMGR PVRQNLAMTG EVSLTGKIL PVGGIKEKTI AAKRAGVTCI VLPAENKKDF YDLAAFITEG LEVHFVEHYR EIFDIAFPDE QAEALAVER UniProtKB: Lon protease homolog, mitochondrial |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 6 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)