[English] 日本語
 Yorodumi
Yorodumi- EMDB-16801: Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein insertion / GET pathway / tail anchored membrane protein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationarsenite transmembrane transporter activity / membrane insertase activity / GET complex / tail-anchored membrane protein insertion into ER membrane / receptor recycling / Hydrolases; Acting on acid anhydrides / protein insertion into ER membrane / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / B cell homeostasis / protein-membrane adaptor activity ...arsenite transmembrane transporter activity / membrane insertase activity / GET complex / tail-anchored membrane protein insertion into ER membrane / receptor recycling / Hydrolases; Acting on acid anhydrides / protein insertion into ER membrane / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / B cell homeostasis / protein-membrane adaptor activity / vesicle-mediated transport / negative regulation of proteasomal ubiquitin-dependent protein catabolic process / negative regulation of protein ubiquitination / epidermal growth factor receptor signaling pathway / defense response / protein stabilization / ubiquitin protein ligase binding / endoplasmic reticulum membrane / nucleolus / signal transduction / endoplasmic reticulum / ATP hydrolysis activity / extracellular exosome / nucleoplasm / ATP binding / membrane / nucleus / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
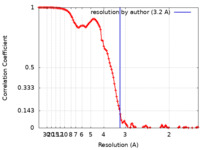
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | McDowell MA / Heimes M / Wild K / Sinning I | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Structural Basis of Tail-Anchored Membrane Protein Biogenesis by the GET Insertase Complex. Authors: Melanie A McDowell / Michael Heimes / Francesco Fiorentino / Shahid Mehmood / Ákos Farkas / Javier Coy-Vergara / Di Wu / Jani Reddy Bolla / Volker Schmid / Roger Heinze / Klemens Wild / ...Authors: Melanie A McDowell / Michael Heimes / Francesco Fiorentino / Shahid Mehmood / Ákos Farkas / Javier Coy-Vergara / Di Wu / Jani Reddy Bolla / Volker Schmid / Roger Heinze / Klemens Wild / Dirk Flemming / Stefan Pfeffer / Blanche Schwappach / Carol V Robinson / Irmgard Sinning /   Abstract: Membrane protein biogenesis faces the challenge of chaperoning hydrophobic transmembrane helices for faithful membrane insertion. The guided entry of tail-anchored proteins (GET) pathway targets and ...Membrane protein biogenesis faces the challenge of chaperoning hydrophobic transmembrane helices for faithful membrane insertion. The guided entry of tail-anchored proteins (GET) pathway targets and inserts tail-anchored (TA) proteins into the endoplasmic reticulum (ER) membrane with an insertase (yeast Get1/Get2 or mammalian WRB/CAML) that captures the TA from a cytoplasmic chaperone (Get3 or TRC40, respectively). Here, we present cryo-electron microscopy reconstructions, native mass spectrometry, and structure-based mutagenesis of human WRB/CAML/TRC40 and yeast Get1/Get2/Get3 complexes. Get3 binding to the membrane insertase supports heterotetramer formation, and phosphatidylinositol binding at the heterotetramer interface stabilizes the insertase for efficient TA insertion in vivo. We identify a Get2/CAML cytoplasmic helix that forms a "gating" interaction with Get3/TRC40 important for TA insertion. Structural homology with YidC and the ER membrane protein complex (EMC) implicates an evolutionarily conserved insertion mechanism for divergent substrates utilizing a hydrophilic groove. Thus, we provide a detailed structural and mechanistic framework to understand TA membrane insertion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16801.map.gz emd_16801.map.gz | 126.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16801-v30.xml emd-16801-v30.xml emd-16801.xml emd-16801.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16801_fsc.xml emd_16801_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_16801.png emd_16801.png | 71 KB | ||
| Masks |  emd_16801_msk_1.map emd_16801_msk_1.map | 134.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16801.cif.gz emd-16801.cif.gz | 6.9 KB | ||
| Others |  emd_16801_additional_1.map.gz emd_16801_additional_1.map.gz emd_16801_half_map_1.map.gz emd_16801_half_map_1.map.gz emd_16801_half_map_2.map.gz emd_16801_half_map_2.map.gz | 125.2 MB 124.6 MB 124.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16801 http://ftp.pdbj.org/pub/emdb/structures/EMD-16801 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16801 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16801 | HTTPS FTP |
-Validation report
| Summary document |  emd_16801_validation.pdf.gz emd_16801_validation.pdf.gz | 770.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16801_full_validation.pdf.gz emd_16801_full_validation.pdf.gz | 770.1 KB | Display | |
| Data in XML |  emd_16801_validation.xml.gz emd_16801_validation.xml.gz | 19.5 KB | Display | |
| Data in CIF |  emd_16801_validation.cif.gz emd_16801_validation.cif.gz | 25.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16801 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16801 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16801 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16801 | HTTPS FTP |
-Related structure data
| Related structure data |  8cr1MC  8cqzC  8cr2C  8oduC  8odvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16801.map.gz / Format: CCP4 / Size: 134.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16801.map.gz / Format: CCP4 / Size: 134.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
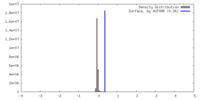
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16801_msk_1.map emd_16801_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
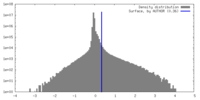
| Density Histograms |
-Additional map: Final reconstruction low-pass filtered to 6 A resolution
| File | emd_16801_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final reconstruction low-pass filtered to 6 A resolution | ||||||||||||
| Projections & Slices |
| ||||||||||||
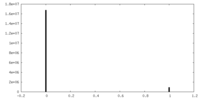
| Density Histograms |
-Half map: #2
| File | emd_16801_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
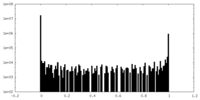
| Density Histograms |
-Half map: #1
| File | emd_16801_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer
| Entire | Name: Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer |
|---|---|
| Components |
|
-Supramolecule #1: Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer
| Supramolecule | Name: Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Get2-Get1 was expressed as a fusion protein in S. frugiperda and Get3 was expressed in E. coli. The complex components were purified and reconstituted in vitro. |
|---|---|
| Molecular weight | Theoretical: 150 KDa |
-Supramolecule #2: Tail-anchored protein insertion receptor Get1 and Get2/CAML
| Supramolecule | Name: Tail-anchored protein insertion receptor Get1 and Get2/CAML type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Dimeric ATPase Get3
| Supramolecule | Name: Dimeric ATPase Get3 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ATPase ASNA1
| Macromolecule | Name: ATPase ASNA1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: Hydrolases; Acting on acid anhydrides |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.14607 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMAAGVAGW GVEAEEFEDA PDVEPLEPTL SNIIEQRSLK WIFVGGKGGV GKTTCSCSLA VQLSKGRESV LIISTDPAHN ISDAFDQKF SKVPTKVKGY DNLFAMEIDP SLGVAELPDE FFEEDNMLSM GKKMMQEAMS AFPGIDEAMS YAEVMRLVKG M NFSVVVFD ...String: GAMAAGVAGW GVEAEEFEDA PDVEPLEPTL SNIIEQRSLK WIFVGGKGGV GKTTCSCSLA VQLSKGRESV LIISTDPAHN ISDAFDQKF SKVPTKVKGY DNLFAMEIDP SLGVAELPDE FFEEDNMLSM GKKMMQEAMS AFPGIDEAMS YAEVMRLVKG M NFSVVVFD TAPTGHTLRL LNFPTIVERG LGRLMQIKNQ ISPFISQMCN MLGLGDMNAD QLASKLEETL PVIRSVSEQF KD PEQTTFI CVCIAEFLSL YETERLIQEL AKCKIDTHNI IVNQLVFPDP EKPCKMCEAR HKIQAKYLDQ MEDLYEDFHI VKL PLLPHE VRGADKVNTF SALLLEPYKP PSAQGSWSHP QFEK UniProtKB: ATPase GET3 |
-Macromolecule #2: Guided entry of tail-anchored proteins factor CAMLG,Guided entry ...
| Macromolecule | Name: Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1,GET2-GET1 type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.083309 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDSFRIFRLV GCALLALGVR AFVCKYLSIF APFLTLQLAY MGLYKYFPKS EKKIKTTVLT AALLLSGIPA EVINRSMDTY SKMGEVFTD LCVYFFTFIF CHELLDYWGS EVPGSGSENL YFQSGSGSMS SAAADHWAWL LVLSFVFGCN VLRILLPSFS S FMSRVLQK ...String: MDSFRIFRLV GCALLALGVR AFVCKYLSIF APFLTLQLAY MGLYKYFPKS EKKIKTTVLT AALLLSGIPA EVINRSMDTY SKMGEVFTD LCVYFFTFIF CHELLDYWGS EVPGSGSENL YFQSGSGSMS SAAADHWAWL LVLSFVFGCN VLRILLPSFS S FMSRVLQK DAEQESQMRA EIQDMKQELS TVNMMDEFAR YARLERKINK MTDKLKTHVK ARTAQLAKIK WVISVAFYVL QA ALMISLI WKYYSVPVAV VPSKWITPLD RLVAFPTRVA GGVGITCWIL VCNKVVAIVL HPFSGSGSLE VLFQ UniProtKB: Guided entry of tail-anchored proteins factor CAMLG, Guided entry of tail-anchored proteins factor 1 |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.8 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER/RHODIUM / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV | ||||||
| Details | Complex stabilised in PMAL-C8 amphipol |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 9470 / Average exposure time: 12.0 sec. / Average electron dose: 45.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8cr1: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)