[English] 日本語
 Yorodumi
Yorodumi- EMDB-16802: Homo sapiens Get1/Get2 heterotetramer (a3' deletion variant) in c... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Homo sapiens Get1/Get2 heterotetramer (a3' deletion variant) in complex with a Get3 dimer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein insertion / GET pathway / tail anchored membrane protein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationarsenite transmembrane transporter activity / otic vesicle development / GET complex / membrane insertase activity / tail-anchored membrane protein insertion into ER membrane / receptor recycling / protein insertion into ER membrane / post-translational protein targeting to endoplasmic reticulum membrane / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / B cell homeostasis ...arsenite transmembrane transporter activity / otic vesicle development / GET complex / membrane insertase activity / tail-anchored membrane protein insertion into ER membrane / receptor recycling / protein insertion into ER membrane / post-translational protein targeting to endoplasmic reticulum membrane / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / B cell homeostasis / vesicle-mediated transport / protein-membrane adaptor activity / negative regulation of protein ubiquitination / negative regulation of proteasomal ubiquitin-dependent protein catabolic process / establishment of localization in cell / sensory perception of sound / defense response / synapse organization / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / epidermal growth factor receptor signaling pathway / protein stabilization / ubiquitin protein ligase binding / endoplasmic reticulum membrane / nucleolus / endoplasmic reticulum / signal transduction / ATP hydrolysis activity / extracellular exosome / nucleoplasm / ATP binding / metal ion binding / membrane / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | McDowell MA / Heimes M / Wild K / Sinning I | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: The GET insertase exhibits conformational plasticity and induces membrane thinning. Authors: Melanie A McDowell / Michael Heimes / Giray Enkavi / Ákos Farkas / Daniel Saar / Klemens Wild / Blanche Schwappach / Ilpo Vattulainen / Irmgard Sinning /   Abstract: The eukaryotic guided entry of tail-anchored proteins (GET) pathway mediates the biogenesis of tail-anchored (TA) membrane proteins at the endoplasmic reticulum. In the cytosol, the Get3 chaperone ...The eukaryotic guided entry of tail-anchored proteins (GET) pathway mediates the biogenesis of tail-anchored (TA) membrane proteins at the endoplasmic reticulum. In the cytosol, the Get3 chaperone captures the TA protein substrate and delivers it to the Get1/Get2 membrane protein complex (GET insertase), which then inserts the substrate via a membrane-embedded hydrophilic groove. Here, we present structures, atomistic simulations and functional data of human and Chaetomium thermophilum Get1/Get2/Get3. The core fold of the GET insertase is conserved throughout eukaryotes, whilst thinning of the lipid bilayer occurs in the vicinity of the hydrophilic groove to presumably lower the energetic barrier of membrane insertion. We show that the gating interaction between Get2 helix α3' and Get3 drives conformational changes in both Get3 and the Get1/Get2 membrane heterotetramer. Thus, we provide a framework to understand the conformational plasticity of the GET insertase and how it remodels its membrane environment to promote substrate insertion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16802.map.gz emd_16802.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16802-v30.xml emd-16802-v30.xml emd-16802.xml emd-16802.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
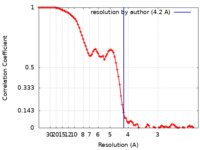
| FSC (resolution estimation) |  emd_16802_fsc.xml emd_16802_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16802.png emd_16802.png | 62.9 KB | ||
| Masks |  emd_16802_msk_1.map emd_16802_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16802.cif.gz emd-16802.cif.gz | 6.8 KB | ||
| Others |  emd_16802_half_map_1.map.gz emd_16802_half_map_1.map.gz emd_16802_half_map_2.map.gz emd_16802_half_map_2.map.gz | 59.3 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16802 http://ftp.pdbj.org/pub/emdb/structures/EMD-16802 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16802 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16802 | HTTPS FTP |
-Related structure data
| Related structure data |  8cr2MC  8cqzC  8cr1C  8oduC  8odvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16802.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16802.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.11 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16802_msk_1.map emd_16802_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16802_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16802_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homo sapiens Get1/Get2 heterotetramer (a3' deletion variant) in c...
| Entire | Name: Homo sapiens Get1/Get2 heterotetramer (a3' deletion variant) in complex with a Get3 dimer |
|---|---|
| Components |
|
-Supramolecule #1: Homo sapiens Get1/Get2 heterotetramer (a3' deletion variant) in c...
| Supramolecule | Name: Homo sapiens Get1/Get2 heterotetramer (a3' deletion variant) in complex with a Get3 dimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Get2-Get1 was expressed as a fusion protein in S. frugiperda and Get3 was expressed in E. coli. The complex components were purified and reconstituted in vitro. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Tail-anchored protein insertion receptor Get1 and Get2/CAML (a3' ...
| Supramolecule | Name: Tail-anchored protein insertion receptor Get1 and Get2/CAML (a3' deletion variant) type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Dimeric ATPase Get3
| Supramolecule | Name: Dimeric ATPase Get3 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ATPase ASNA1
| Macromolecule | Name: ATPase ASNA1 / type: protein_or_peptide / ID: 1 / Details: Get3 was expressed in E. coli / Number of copies: 2 / Enantiomer: LEVO / EC number: Hydrolases; Acting on acid anhydrides |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.14607 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMAAGVAGW GVEAEEFEDA PDVEPLEPTL SNIIEQRSLK WIFVGGKGGV GKTTCSCSLA VQLSKGRESV LIISTDPAHN ISDAFDQKF SKVPTKVKGY DNLFAMEIDP SLGVAELPDE FFEEDNMLSM GKKMMQEAMS AFPGIDEAMS YAEVMRLVKG M NFSVVVFD ...String: GAMAAGVAGW GVEAEEFEDA PDVEPLEPTL SNIIEQRSLK WIFVGGKGGV GKTTCSCSLA VQLSKGRESV LIISTDPAHN ISDAFDQKF SKVPTKVKGY DNLFAMEIDP SLGVAELPDE FFEEDNMLSM GKKMMQEAMS AFPGIDEAMS YAEVMRLVKG M NFSVVVFD TAPTGHTLRL LNFPTIVERG LGRLMQIKNQ ISPFISQMCN MLGLGDMNAD QLASKLEETL PVIRSVSEQF KD PEQTTFI CVCIAEFLSL YETERLIQEL AKCKIDTHNI IVNQLVFPDP EKPCKMCEAR HKIQAKYLDQ MEDLYEDFHI VKL PLLPHE VRGADKVNTF SALLLEPYKP PSAQGSWSHP QFEK UniProtKB: ATPase GET3 |
-Macromolecule #2: Guided entry of tail-anchored proteins factor CAMLG,Guided entry ...
| Macromolecule | Name: Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1 type: protein_or_peptide / ID: 2 Details: Get2-Get1 was expressed as a fusion protein in S. frugiperda Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.429418 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDSFRIFRLV GCALLALGVR AFVCKYLSIF APFLTLQLAY MGLYKYFPKS EKKIKTTGGG GGIPAEVINR SMDTYSKMGE VFTDLCVYF FTFIFCHELL DYWGSEVPGS GSENLYFQSG SGSMSSAAAD HWAWLLVLSF VFGCNVLRIL LPSFSSFMSR V LQKDAEQE ...String: MDSFRIFRLV GCALLALGVR AFVCKYLSIF APFLTLQLAY MGLYKYFPKS EKKIKTTGGG GGIPAEVINR SMDTYSKMGE VFTDLCVYF FTFIFCHELL DYWGSEVPGS GSENLYFQSG SGSMSSAAAD HWAWLLVLSF VFGCNVLRIL LPSFSSFMSR V LQKDAEQE SQMRAEIQDM KQELSTVNMM DEFARYARLE RKINKMTDKL KTHVKARTAQ LAKIKWVISV AFYVLQAALM IS LIWKYYS VPVAVVPSKW ITPLDRLVAF PTRVAGGVGI TCWILVCNKV VAIVLHPFSG SGSLEVLFQ UniProtKB: Guided entry of tail-anchored proteins factor CAMLG, Guided entry of tail-anchored proteins factor CAMLG, Guided entry of tail-anchored proteins factor 1 |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV | ||||||
| Details | Complex stabilised in PMAL-C8 amphipol |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 12311 / Average exposure time: 2.58 sec. / Average electron dose: 53.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)