+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-1669 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structures of the idle yeast Ssh1 complex bound to the yeast 80S ribosome | |||||||||
 マップデータ マップデータ | This map represents a yeast 80S ribosome bound to the idle yeast Ssh1 complex | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Ribosome / protein exit tunnel / cotranslational protein translocation / protein conducting channel / signal sequence | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報: / translocon complex / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / cytoplasmic translational elongation / rough endoplasmic reticulum membrane / Ssh1 translocon complex / Sec61 translocon complex / SRP-dependent cotranslational protein targeting to membrane, translocation / signal sequence binding / SRP-dependent cotranslational protein targeting to membrane ...: / translocon complex / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / cytoplasmic translational elongation / rough endoplasmic reticulum membrane / Ssh1 translocon complex / Sec61 translocon complex / SRP-dependent cotranslational protein targeting to membrane, translocation / signal sequence binding / SRP-dependent cotranslational protein targeting to membrane / post-translational protein targeting to membrane, translocation / regulation of amino acid metabolic process / negative regulation of glucose mediated signaling pathway / positive regulation of translational fidelity / RMTs methylate histone arginines / Protein methylation / nuclear inner membrane / Protein hydroxylation / ribosome-associated ubiquitin-dependent protein catabolic process / pre-mRNA 5'-splice site binding / GDP-dissociation inhibitor activity / positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / nonfunctional rRNA decay / Formation of the ternary complex, and subsequently, the 43S complex / Translation initiation complex formation / response to cycloheximide / Ribosomal scanning and start codon recognition / cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / preribosome, small subunit precursor / Major pathway of rRNA processing in the nucleolus and cytosol / mRNA destabilization / SRP-dependent cotranslational protein targeting to membrane / GTP hydrolysis and joining of the 60S ribosomal subunit / negative regulation of mRNA splicing, via spliceosome / preribosome, large subunit precursor / Formation of a pool of free 40S subunits / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / negative regulation of translational frameshifting / L13a-mediated translational silencing of Ceruloplasmin expression / endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / protein transmembrane transporter activity / translational elongation / G-protein alpha-subunit binding / ribosomal large subunit export from nucleus / protein kinase activator activity / 90S preribosome / ribonucleoprotein complex binding / translational termination / regulation of translational fidelity / ribosomal subunit export from nucleus / protein-RNA complex assembly / maturation of LSU-rRNA / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / translation regulator activity / ribosomal small subunit export from nucleus / DNA-(apurinic or apyrimidinic site) endonuclease activity / rescue of stalled cytosolic ribosome / protein kinase C binding / guanyl-nucleotide exchange factor activity / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / macroautophagy / maturation of SSU-rRNA / translational initiation / small-subunit processome / maintenance of translational fidelity / modification-dependent protein catabolic process / protein tag activity / cytoplasmic stress granule / rRNA processing / large ribosomal subunit / ribosome biogenesis / ribosome binding / ribosomal small subunit biogenesis / ribosomal small subunit assembly / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / negative regulation of translation / rRNA binding / protein ubiquitination / structural constituent of ribosome / ribosome / translation / G protein-coupled receptor signaling pathway / negative regulation of gene expression / response to antibiotic / mRNA binding / ubiquitin protein ligase binding / endoplasmic reticulum membrane / nucleolus / structural molecule activity / endoplasmic reticulum 類似検索 - 分子機能 | |||||||||
| 生物種 |  | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / ネガティブ染色法 / 解像度: 8.8 Å | |||||||||
 データ登録者 データ登録者 | Becker T / Mandon E / Bhushan S / Jarasch A / Armache JP / Funes S / Jossinet F / Gumbart J / Mielke T / Berninghausen O ...Becker T / Mandon E / Bhushan S / Jarasch A / Armache JP / Funes S / Jossinet F / Gumbart J / Mielke T / Berninghausen O / Schulten K / Westhof E / Gilmore R / Beckmann R | |||||||||
 引用 引用 |  ジャーナル: Science / 年: 2009 ジャーナル: Science / 年: 2009タイトル: Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. 著者: Thomas Becker / Shashi Bhushan / Alexander Jarasch / Jean-Paul Armache / Soledad Funes / Fabrice Jossinet / James Gumbart / Thorsten Mielke / Otto Berninghausen / Klaus Schulten / Eric ...著者: Thomas Becker / Shashi Bhushan / Alexander Jarasch / Jean-Paul Armache / Soledad Funes / Fabrice Jossinet / James Gumbart / Thorsten Mielke / Otto Berninghausen / Klaus Schulten / Eric Westhof / Reid Gilmore / Elisabet C Mandon / Roland Beckmann /  要旨: The trimeric Sec61/SecY complex is a protein-conducting channel (PCC) for secretory and membrane proteins. Although Sec complexes can form oligomers, it has been suggested that a single copy may ...The trimeric Sec61/SecY complex is a protein-conducting channel (PCC) for secretory and membrane proteins. Although Sec complexes can form oligomers, it has been suggested that a single copy may serve as an active PCC. We determined subnanometer-resolution cryo-electron microscopy structures of eukaryotic ribosome-Sec61 complexes. In combination with biochemical data, we found that in both idle and active states, the Sec complex is not oligomeric and interacts mainly via two cytoplasmic loops with the universal ribosomal adaptor site. In the active state, the ribosomal tunnel and a central pore of the monomeric PCC were occupied by the nascent chain, contacting loop 6 of the Sec complex. This provides a structural basis for the activity of a solitary Sec complex in cotranslational protein translocation. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_1669.map.gz emd_1669.map.gz | 21.5 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-1669-v30.xml emd-1669-v30.xml emd-1669.xml emd-1669.xml | 12.4 KB 12.4 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  1669.gif 1669.gif 1669_EMD_1669_inactive_yeast.jpg 1669_EMD_1669_inactive_yeast.jpg | 91.2 KB 220.3 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1669 http://ftp.pdbj.org/pub/emdb/structures/EMD-1669 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1669 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1669 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_1669_validation.pdf.gz emd_1669_validation.pdf.gz | 356.4 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_1669_full_validation.pdf.gz emd_1669_full_validation.pdf.gz | 356 KB | 表示 | |
| XML形式データ |  emd_1669_validation.xml.gz emd_1669_validation.xml.gz | 7.2 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1669 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1669 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1669 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1669 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_1669.map.gz / 形式: CCP4 / 大きさ: 185.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_1669.map.gz / 形式: CCP4 / 大きさ: 185.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | This map represents a yeast 80S ribosome bound to the idle yeast Ssh1 complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.2375 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
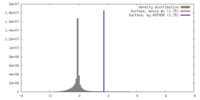
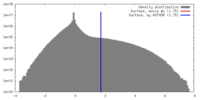
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : An idle yeast Ssh1 complex bound to a yeast 80S ribosome
| 全体 | 名称: An idle yeast Ssh1 complex bound to a yeast 80S ribosome |
|---|---|
| 要素 |
|
-超分子 #1000: An idle yeast Ssh1 complex bound to a yeast 80S ribosome
| 超分子 | 名称: An idle yeast Ssh1 complex bound to a yeast 80S ribosome タイプ: sample / ID: 1000 詳細: 80S ribosomes and the detergent solubilized Ssh1 complex were reconstituted in vitro by adding 1 pmol of ribosome and Ssh1 complex in 5 fold molar excess 集合状態: 80S Ribosome bound to one copy of the heterotrimeric Ssh1 complex Number unique components: 2 |
|---|---|
| 分子量 | 実験値: 4.2 MDa / 理論値: 4.2 MDa / 手法: Known for 80S ribosomes |
-超分子 #1: Yeast 80S ribosome bound to the yeast Ssh1 complex
| 超分子 | 名称: Yeast 80S ribosome bound to the yeast Ssh1 complex / タイプ: complex / ID: 1 Name.synonym: Yeast 80S ribosome bound to the yeast Ssh1 complex Ribosome-details: ribosome-eukaryote: ALL |
|---|---|
| 分子量 | 実験値: 4.2 MDa / 理論値: 4.2 MDa |
-分子 #1: Ssh1 complex
| 分子 | 名称: Ssh1 complex / タイプ: ligand / ID: 1 / Name.synonym: Ssh1 complex / 詳細: His FLAG-tagged / コピー数: 1 / 集合状態: Heterotrimer / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 71.5 KDa |
| 組換発現 | 生物種:  |
-実験情報
-構造解析
| 手法 | ネガティブ染色法, クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 詳細: 20 mM HEPES/KOH, pH 7.5 100 mM KOAc, 10 mM Mg(OAc)2, 1.5 mM DTT, 0.1 % (w/v) digitonin |
|---|---|
| 染色 | タイプ: NEGATIVE / 詳細: Cryo-EM |
| グリッド | 詳細: Quantifoil grids (3/3) with 2 nm carbon on top |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 95 % / 装置: OTHER / 詳細: Vitrification instrument: Vitrobot 手法: Blot for 10 seconds before plunging, use 2 layer of filter paper |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI POLARA 300 |
|---|---|
| 温度 | 平均: 84 K |
| アライメント法 | Legacy - 非点収差: Objective lens astigmatism was corrected at 100000 times magnification |
| 撮影 | カテゴリ: FILM / フィルム・検出器のモデル: KODAK SO-163 FILM / デジタル化 - スキャナー: PRIMESCAN / デジタル化 - サンプリング間隔: 4.76 µm / 実像数: 185 / 平均電子線量: 25 e/Å2 / 詳細: Scanned at 5334 dpi / Od range: 1.2 / ビット/ピクセル: 16 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 倍率(補正後): 38000 / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.26 mm / 最大 デフォーカス(公称値): 4.5 µm / 最小 デフォーカス(公称値): 1.2 µm / 倍率(公称値): 39000 |
| 試料ステージ | 試料ホルダー: FEI Polara cartridge system / 試料ホルダーモデル: OTHER |
| 実験機器 |  モデル: Tecnai Polara / 画像提供: FEI Company |
- 画像解析
画像解析
| 詳細 | Particles were selected using the program SIGNATURE and visually inspected. This map resulted from sorting against the ES27 exit position and subsequent sorting for tRNA and the Ssh1 complex and represents the datasubset with the Ssh1 complex present without tRNA. |
|---|---|
| CTF補正 | 詳細: Defocus group volumes |
| 最終 再構成 | 想定した対称性 - 点群: C1 (非対称) / アルゴリズム: OTHER / 解像度のタイプ: BY AUTHOR / 解像度: 8.8 Å / 解像度の算出法: FSC 0.5 CUT-OFF / ソフトウェア - 名称: SPIDER 詳細: Map was filtered between 8.3 and 10.3 Angstrom to better visualize the Ssh1 complex 使用した粒子像数: 20400 |
| 最終 角度割当 | 詳細: SPIDER |
 ムービー
ムービー コントローラー
コントローラー



































 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)