+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C2) - consensus map | ||||||||||||
 マップデータ マップデータ | |||||||||||||
 試料 試料 |
| ||||||||||||
| 生物種 |  | ||||||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.9 Å | ||||||||||||
 データ登録者 データ登録者 | Best KM / Ikeuchi K / Kater L / Best DM / Musial J / Matsuo Y / Berninghausen O / Becker T / Inada T / Beckmann R | ||||||||||||
| 資金援助 | European Union,  ドイツ, 3件 ドイツ, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2023 ジャーナル: Nat Commun / 年: 2023タイトル: Structural basis for clearing of ribosome collisions by the RQT complex. 著者: Katharina Best / Ken Ikeuchi / Lukas Kater / Daniel Best / Joanna Musial / Yoshitaka Matsuo / Otto Berninghausen / Thomas Becker / Toshifumi Inada / Roland Beckmann /    要旨: Translation of aberrant messenger RNAs can cause stalling of ribosomes resulting in ribosomal collisions. Collided ribosomes are specifically recognized to initiate stress responses and quality ...Translation of aberrant messenger RNAs can cause stalling of ribosomes resulting in ribosomal collisions. Collided ribosomes are specifically recognized to initiate stress responses and quality control pathways. Ribosome-associated quality control facilitates the degradation of incomplete translation products and requires dissociation of the stalled ribosomes. A central event is therefore the splitting of collided ribosomes by the ribosome quality control trigger complex, RQT, by an unknown mechanism. Here we show that RQT requires accessible mRNA and the presence of a neighboring ribosome. Cryogenic electron microscopy of RQT-ribosome complexes reveals that RQT engages the 40S subunit of the lead ribosome and can switch between two conformations. We propose that the Ski2-like helicase 1 (Slh1) subunit of RQT applies a pulling force on the mRNA, causing destabilizing conformational changes of the small ribosomal subunit, ultimately resulting in subunit dissociation. Our findings provide conceptual framework for a helicase-driven ribosomal splitting mechanism. #1:  ジャーナル: Acta Crystallogr D Struct Biol / 年: 2018 ジャーナル: Acta Crystallogr D Struct Biol / 年: 2018タイトル: Real-space refinement in PHENIX for cryo-EM and crystallography. 著者: Afonine PV / Poon BK / Read RJ / Sobolev OV / Terwilliger TC / Urzhumtsev A / Adams PD | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_16571.map.gz emd_16571.map.gz | 336 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-16571-v30.xml emd-16571-v30.xml emd-16571.xml emd-16571.xml | 37.1 KB 37.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_16571_fsc.xml emd_16571_fsc.xml | 18.4 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_16571.png emd_16571.png | 139.3 KB | ||
| マスクデータ |  emd_16571_msk_1.map emd_16571_msk_1.map | 669.9 MB |  マスクマップ マスクマップ | |
| その他 |  emd_16571_half_map_1.map.gz emd_16571_half_map_1.map.gz emd_16571_half_map_2.map.gz emd_16571_half_map_2.map.gz | 621.6 MB 621.5 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16571 http://ftp.pdbj.org/pub/emdb/structures/EMD-16571 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16571 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16571 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_16571_validation.pdf.gz emd_16571_validation.pdf.gz | 1 MB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_16571_full_validation.pdf.gz emd_16571_full_validation.pdf.gz | 1 MB | 表示 | |
| XML形式データ |  emd_16571_validation.xml.gz emd_16571_validation.xml.gz | 28.6 KB | 表示 | |
| CIF形式データ |  emd_16571_validation.cif.gz emd_16571_validation.cif.gz | 37.9 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16571 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16571 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16571 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16571 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_16571.map.gz / 形式: CCP4 / 大きさ: 669.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_16571.map.gz / 形式: CCP4 / 大きさ: 669.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
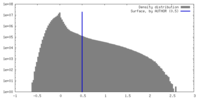
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_16571_msk_1.map emd_16571_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
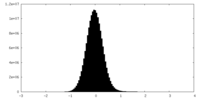
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_16571_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_16571_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : ribosome with bound RQT components (Slh1, Cue3 and Rqt4)
| 全体 | 名称: ribosome with bound RQT components (Slh1, Cue3 and Rqt4) |
|---|---|
| 要素 |
|
-超分子 #1: ribosome with bound RQT components (Slh1, Cue3 and Rqt4)
| 超分子 | 名称: ribosome with bound RQT components (Slh1, Cue3 and Rqt4) タイプ: complex / ID: 1 / キメラ: Yes / 親要素: 0 / 含まれる分子: #1-#82 |
|---|---|
| 由来(天然) | 生物種:  |
-超分子 #2: RQT complex (Slh1, Cue3 and Rqt4)
| 超分子 | 名称: RQT complex (Slh1, Cue3 and Rqt4) / タイプ: complex / ID: 2 / キメラ: Yes / 親要素: 1 / 含まれる分子: #80-#82 |
|---|---|
| 由来(天然) | 生物種:  |
-超分子 #3: ribosome
| 超分子 | 名称: ribosome / タイプ: complex / ID: 3 / キメラ: Yes / 親要素: 1 / 含まれる分子: #1-#79 |
|---|---|
| 由来(天然) | 生物種:  |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| グリッド | モデル: Quantifoil R3/3 / 材質: COPPER / 支持フィルム - 材質: CARBON |
| 凍結 | 凍結剤: ETHANE / 装置: FEI VITROBOT MARK II |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / 平均電子線量: 43.6 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: SPOT SCAN / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 3.0 µm / 最小 デフォーカス(公称値): 0.5 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)