+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human apo TRPM8 in a closed state (consensus map) | |||||||||
 Map data Map data | map sharpened locally with Local Filtering tool in cryoSPARC | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | calcium ion channel / cold temperature sensor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationligand-gated calcium channel activity / thermoception / TRP channels / response to cold / calcium ion transmembrane transport / calcium channel activity / intracellular calcium ion homeostasis / positive regulation of cold-induced thermogenesis / membrane raft / external side of plasma membrane ...ligand-gated calcium channel activity / thermoception / TRP channels / response to cold / calcium ion transmembrane transport / calcium channel activity / intracellular calcium ion homeostasis / positive regulation of cold-induced thermogenesis / membrane raft / external side of plasma membrane / endoplasmic reticulum membrane / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.65 Å | |||||||||
 Authors Authors | Palchevskyi S / Czarnocki-Cieciura M / Vistoli G / Gervasoni S / Nowak E / Beccari AR / Nowotny M / Talarico C | |||||||||
| Funding support |  Poland, 1 items Poland, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Structure of human TRPM8 channel. Authors: Sergii Palchevskyi / Mariusz Czarnocki-Cieciura / Giulio Vistoli / Silvia Gervasoni / Elżbieta Nowak / Andrea R Beccari / Marcin Nowotny / Carmine Talarico /    Abstract: TRPM8 is a non-selective cation channel permeable to both monovalent and divalent cations that is activated by multiple factors, such as temperature, voltage, pressure, and changes in osmolality. It ...TRPM8 is a non-selective cation channel permeable to both monovalent and divalent cations that is activated by multiple factors, such as temperature, voltage, pressure, and changes in osmolality. It is a therapeutic target for anticancer drug development, and its modulators can be utilized for several pathological conditions. Here, we present a cryo-electron microscopy structure of a human TRPM8 channel in the closed state that was solved at 2.7 Å resolution. Our structure comprises the most complete model of the N-terminal pre-melastatin homology region. We also visualized several lipids that are bound by the protein and modeled how the human channel interacts with icilin. Analyses of pore helices in available TRPM structures showed that all these structures can be grouped into different closed, desensitized and open state conformations based on the register of the pore helix S6 which positions particular amino acid residues at the channel constriction. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15982.map.gz emd_15982.map.gz | 13.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15982-v30.xml emd-15982-v30.xml emd-15982.xml emd-15982.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15982_fsc.xml emd_15982_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_15982.png emd_15982.png | 144.9 KB | ||
| Masks |  emd_15982_msk_1.map emd_15982_msk_1.map | 282.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15982.cif.gz emd-15982.cif.gz | 6.1 KB | ||
| Others |  emd_15982_additional_1.map.gz emd_15982_additional_1.map.gz emd_15982_half_map_1.map.gz emd_15982_half_map_1.map.gz emd_15982_half_map_2.map.gz emd_15982_half_map_2.map.gz | 141.1 MB 261.8 MB 261.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15982 http://ftp.pdbj.org/pub/emdb/structures/EMD-15982 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15982 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15982 | HTTPS FTP |
-Validation report
| Summary document |  emd_15982_validation.pdf.gz emd_15982_validation.pdf.gz | 870.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15982_full_validation.pdf.gz emd_15982_full_validation.pdf.gz | 870.3 KB | Display | |
| Data in XML |  emd_15982_validation.xml.gz emd_15982_validation.xml.gz | 22.8 KB | Display | |
| Data in CIF |  emd_15982_validation.cif.gz emd_15982_validation.cif.gz | 29.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15982 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15982 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15982 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15982 | HTTPS FTP |
-Related structure data
| Related structure data |  8bdcC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15982.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15982.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map sharpened locally with Local Filtering tool in cryoSPARC | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15982_msk_1.map emd_15982_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
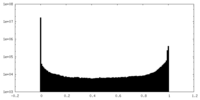
| Density Histograms |
-Additional map: raw map
| File | emd_15982_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | raw map | ||||||||||||
| Projections & Slices |
| ||||||||||||
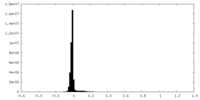
| Density Histograms |
-Half map: half map A
| File | emd_15982_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
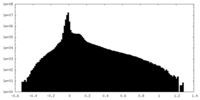
| Density Histograms |
-Half map: half map B
| File | emd_15982_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
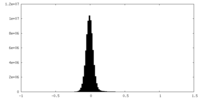
| Density Histograms |
- Sample components
Sample components
-Entire : Human TRPM8 in apo form
| Entire | Name: Human TRPM8 in apo form |
|---|---|
| Components |
|
-Supramolecule #1: Human TRPM8 in apo form
| Supramolecule | Name: Human TRPM8 in apo form / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 510 KDa |
-Macromolecule #1: TRPM8
| Macromolecule | Name: TRPM8 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: SNSFRAARLS MRNRRNDTLD STRTLYSSAS RSTDLSYSES DLVNFIQANF KKRECVFFTK DSKATENVCK CGYAQSQHME GTQINQSEKW NYKKHTKEFP TDAFGDIQFE TLGKKGKYIR LSCDTDAEIL YELLTQHWHL KTPNLVISVT GGAKNFALKP RMRKIFSRLI ...String: SNSFRAARLS MRNRRNDTLD STRTLYSSAS RSTDLSYSES DLVNFIQANF KKRECVFFTK DSKATENVCK CGYAQSQHME GTQINQSEKW NYKKHTKEFP TDAFGDIQFE TLGKKGKYIR LSCDTDAEIL YELLTQHWHL KTPNLVISVT GGAKNFALKP RMRKIFSRLI YIAQSKGAWI LTGGTHYGLM KYIGEVVRDN TISRSSEENI VAIGIAAWGM VSNRDTLIRN CDAEGYFLAQ YLMDDFTRDP LYILDNNHTH LLLVDNGCHG HPTVEAKLRN QLEKYISERT IQDSNYGGKI PIVCFAQGGG KETLKAINTS IKNKIPCVVV EGSGQIADVI ASLVEVEDAL TSSAVKEKLV RFLPRTVSRL PEEETESWIK WLKEILECSH LLTVIKMEEA GDEIVSNAIS YALYKAFSTS EQDKDNWNGQ LKLLLEWNQL DLANDEIFTN DRRWESADLQ EVMFTALIKD RPKFVRLFLE NGLNLRKFLT HDVLTELFSN HFSTLVYRNL QIAKNSYNDA LLTFVWKLVA NFRRGFRKED RNGRDEMDIE LHDVSPITRH PLQALFIWAI LQNKKELSKV IWEQTRGCTL AALGASKLLK TLAKVKNDIN AAGESEELAN EYETRAVELF TECYSSDEDL AEQLLVYSCE AWGGSNCLEL AVEATDQHFI AQPGVQNFLS KQWYGEISRD TKNWKIILCL FIIPLVGCGF VSFRKKPVDK HKKLLWYYVA FFTSPFVVFS WNVVFYIAFL LLFAYVLLMD FHSVPHPPEL VLYSLVFVLF CDEVRQWYVN GVNYFTDLWN VMDTLGLFYF IAGIVFRLHS SNKSSLYSGR VIFCLDYIIF TLRLIHIFTV SRNLGPKIIM LQRMLIDVFF FLFLFAVWMV AFGVARQGIL RQNEQRWRWI FRSVIYEPYL AMFGQVPSDV DGTTYDFAHC TFTGNESKPL CVELDEHNLP RFPEWITIPL VCIYMLSTNI LLVNLLVAMF GYTVGTVQEN NDQVWKFQRY FLVQEYCSRL NIPFPFIVFA YFYMVVKKCF KCCCKEKNME SSVCCFKNED NETLAWEGVM KENYLVKINT KANDTSEEMR HRFRQLDTKL NDLKGLLKEI ANKIK UniProtKB: Transient receptor potential cation channel subfamily M member 8 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE Details: The grid was glow-discharged from both sides prior to vitrification |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | protein solubilized by LMNG |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 7918 / Average electron dose: 41.42 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)