+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Human Mre11-Nbs1 complex | |||||||||||||||
 マップデータ マップデータ | map_sharp | |||||||||||||||
 試料 試料 |
| |||||||||||||||
 キーワード キーワード | DNA repair / complex / HYDROLASE | |||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報telomere maintenance via telomere trimming / chromosomal region / telomeric 3' overhang formation / mitochondrial double-strand break repair via homologous recombination / Mre11 complex / negative regulation of double-strand break repair via nonhomologous end joining / blastocyst growth / telomere maintenance in response to DNA damage / negative regulation of telomere capping / protection from non-homologous end joining at telomere ...telomere maintenance via telomere trimming / chromosomal region / telomeric 3' overhang formation / mitochondrial double-strand break repair via homologous recombination / Mre11 complex / negative regulation of double-strand break repair via nonhomologous end joining / blastocyst growth / telomere maintenance in response to DNA damage / negative regulation of telomere capping / protection from non-homologous end joining at telomere / meiotic DNA double-strand break formation / Sensing of DNA Double Strand Breaks / BRCA1-C complex / regulation of mitotic recombination / R-loop processing / phosphorylation-dependent protein binding / t-circle formation / chromatin-protein adaptor activity / homologous chromosome pairing at meiosis / DNA strand resection involved in replication fork processing / DNA double-strand break processing / nuclease activity / single-stranded DNA endodeoxyribonuclease activity / homologous recombination / nuclear inclusion body / positive regulation of telomere maintenance / Cytosolic sensors of pathogen-associated DNA / isotype switching / protein localization to site of double-strand break / Impaired BRCA2 binding to PALB2 / HDR through MMEJ (alt-NHEJ) / IRF3-mediated induction of type I IFN / mitotic G2/M transition checkpoint / positive regulation of kinase activity / reciprocal meiotic recombination / sister chromatid cohesion / mitotic intra-S DNA damage checkpoint signaling / Defective homologous recombination repair (HRR) due to BRCA1 loss of function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA1 binding function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA2/RAD51/RAD51C binding function / Homologous DNA Pairing and Strand Exchange / Resolution of D-loop Structures through Synthesis-Dependent Strand Annealing (SDSA) / regulation of DNA-templated DNA replication initiation / Resolution of D-loop Structures through Holliday Junction Intermediates / DNA duplex unwinding / 3'-5'-DNA exonuclease activity / HDR through Single Strand Annealing (SSA) / Impaired BRCA2 binding to RAD51 / double-strand break repair via alternative nonhomologous end joining / mitotic G2 DNA damage checkpoint signaling / Presynaptic phase of homologous DNA pairing and strand exchange / protein K63-linked ubiquitination / neuromuscular process controlling balance / positive regulation of double-strand break repair via homologous recombination / DNA damage response, signal transduction by p53 class mediator / telomere maintenance via telomerase / neuroblast proliferation / positive regulation of protein autophosphorylation / 3'-5' exonuclease activity / telomere maintenance / protein serine/threonine kinase activator activity / intrinsic apoptotic signaling pathway / meiotic cell cycle / DNA endonuclease activity / replication fork / DNA damage checkpoint signaling / Nonhomologous End-Joining (NHEJ) / double-strand break repair via homologous recombination / G2/M DNA damage checkpoint / HDR through Homologous Recombination (HRR) / DNA Damage/Telomere Stress Induced Senescence / PML body / Meiotic recombination / double-strand break repair via nonhomologous end joining / double-strand break repair / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / site of double-strand break / manganese ion binding / Processing of DNA double-strand break ends / histone binding / double-stranded DNA binding / DNA recombination / DNA-binding transcription factor binding / Regulation of TP53 Activity through Phosphorylation / cell population proliferation / chromosome, telomeric region / damaged DNA binding / 加水分解酵素; エステル加水分解酵素 / regulation of cell cycle / cadherin binding / DNA repair / DNA damage response / nucleolus / negative regulation of apoptotic process / Golgi apparatus / nucleoplasm / identical protein binding / nucleus / cytoplasm / cytosol 類似検索 - 分子機能 | |||||||||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||||||||
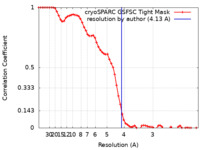
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.13 Å | |||||||||||||||
 データ登録者 データ登録者 | Rotheneder M / Stakyte K / Bartho JD / Lammens K / Hopfner KP | |||||||||||||||
| 資金援助 |  ドイツ, 4件 ドイツ, 4件
| |||||||||||||||
 引用 引用 |  ジャーナル: Mol Cell / 年: 2023 ジャーナル: Mol Cell / 年: 2023タイトル: Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions. 著者: Matthias Rotheneder / Kristina Stakyte / Erik van de Logt / Joseph D Bartho / Katja Lammens / Yilan Fan / Aaron Alt / Brigitte Kessler / Christophe Jung / Wynand P Roos / Barbara ...著者: Matthias Rotheneder / Kristina Stakyte / Erik van de Logt / Joseph D Bartho / Katja Lammens / Yilan Fan / Aaron Alt / Brigitte Kessler / Christophe Jung / Wynand P Roos / Barbara Steigenberger / Karl-Peter Hopfner /  要旨: The DNA double-strand break repair complex Mre11-Rad50-Nbs1 (MRN) detects and nucleolytically processes DNA ends, activates the ATM kinase, and tethers DNA at break sites. How MRN can act both as ...The DNA double-strand break repair complex Mre11-Rad50-Nbs1 (MRN) detects and nucleolytically processes DNA ends, activates the ATM kinase, and tethers DNA at break sites. How MRN can act both as nuclease and scaffold protein is not well understood. The cryo-EM structure of MRN from Chaetomium thermophilum reveals a 2:2:1 complex with a single Nbs1 wrapping around the autoinhibited Mre11 nuclease dimer. MRN has two DNA-binding modes, one ATP-dependent mode for loading onto DNA ends and one ATP-independent mode through Mre11's C terminus, suggesting how it may interact with DSBs and intact DNA. MRNs two 60-nm-long coiled-coil domains form a linear rod structure, the apex of which is assembled by the two joined zinc-hook motifs. Apices from two MRN complexes can further dimerize, forming 120-nm spanning MRN-MRN structures. Our results illustrate the architecture of MRN and suggest how it mechanistically integrates catalytic and tethering functions. | |||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
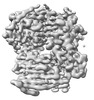
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_15948.map.gz emd_15948.map.gz | 20.9 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-15948-v30.xml emd-15948-v30.xml emd-15948.xml emd-15948.xml | 26.1 KB 26.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_15948_fsc.xml emd_15948_fsc.xml | 6.8 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_15948.png emd_15948.png | 65.6 KB | ||
| マスクデータ |  emd_15948_msk_1.map emd_15948_msk_1.map | 22.2 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-15948.cif.gz emd-15948.cif.gz | 7.4 KB | ||
| その他 |  emd_15948_additional_1.map.gz emd_15948_additional_1.map.gz emd_15948_additional_2.map.gz emd_15948_additional_2.map.gz emd_15948_half_map_1.map.gz emd_15948_half_map_1.map.gz emd_15948_half_map_2.map.gz emd_15948_half_map_2.map.gz | 10.9 MB 169.7 MB 20.6 MB 20.6 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15948 http://ftp.pdbj.org/pub/emdb/structures/EMD-15948 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15948 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15948 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_15948_validation.pdf.gz emd_15948_validation.pdf.gz | 768 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_15948_full_validation.pdf.gz emd_15948_full_validation.pdf.gz | 767.6 KB | 表示 | |
| XML形式データ |  emd_15948_validation.xml.gz emd_15948_validation.xml.gz | 13.3 KB | 表示 | |
| CIF形式データ |  emd_15948_validation.cif.gz emd_15948_validation.cif.gz | 17 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15948 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15948 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15948 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15948 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8bahMC  7zqyC  7zr1C M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_15948.map.gz / 形式: CCP4 / 大きさ: 22.2 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_15948.map.gz / 形式: CCP4 / 大きさ: 22.2 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | map_sharp | ||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.059 Å | ||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
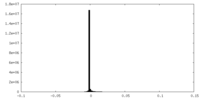
| ファイル |  emd_15948_msk_1.map emd_15948_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-追加マップ: Map unsharpened
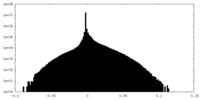
| ファイル | emd_15948_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Map_unsharpened | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-追加マップ: LAFTER Filtered map
| ファイル | emd_15948_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | LAFTER_Filtered_map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Half map B
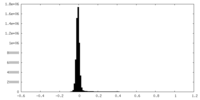
| ファイル | emd_15948_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half_map_B | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Half map A
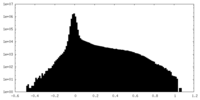
| ファイル | emd_15948_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half_map_A | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Human Mre11-Nbs1 complex
| 全体 | 名称: Human Mre11-Nbs1 complex |
|---|---|
| 要素 |
|
-超分子 #1: Human Mre11-Nbs1 complex
| 超分子 | 名称: Human Mre11-Nbs1 complex / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#2 / 詳細: Human Mre11-dimer with Nbs1 C-terminal chain bound. |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Double-strand break repair protein MRE11
| 分子 | 名称: Double-strand break repair protein MRE11 / タイプ: protein_or_peptide / ID: 1 / コピー数: 2 / 光学異性体: LEVO EC番号: 加水分解酵素; エステル加水分解酵素 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 84.008633 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: MSTADALDDE NTFKILVATD IHLGFMEKDA VRGNDTFVTL DEILRLAQEN EVDFILLGGD LFHENKPSRK TLHTCLELLR KYCMGDRPV QFEILSDQSV NFGFSKFPWV NYQDGNLNIS IPVFSIHGNN DDPTGADALC ALDILSCAGF VNHFGRSMSV E KIDISPVL ...文字列: MSTADALDDE NTFKILVATD IHLGFMEKDA VRGNDTFVTL DEILRLAQEN EVDFILLGGD LFHENKPSRK TLHTCLELLR KYCMGDRPV QFEILSDQSV NFGFSKFPWV NYQDGNLNIS IPVFSIHGNN DDPTGADALC ALDILSCAGF VNHFGRSMSV E KIDISPVL LQKGSTKIAL YGLGSIPDER LYRMFVNKKV TMLRPKEDEN SWFNLFVIHQ NRSKHGSTNF IPEQFLDDFI DL VIWGHEH ECKIAPTKNE QQLFYISQPG SSVVTSLSPG EAVKKHVGLL RIKGRKMNMH KIPLHTVRQF FMEDIVLANH PDI FNPDNP KVTQAIQSFC LEKIEEMLEN AERERLGNSH QPEKPLVRLR VDYSGGFEPF SVLRFSQKFV DRVANPKDII HFFR HREQK EKTGEEINFG KLITKPSEGT TLRVEDLVKQ YFQTAEKNVQ LSLLTERGMG EAVQEFVDKE EKDAIEELVK YQLEK TQRF LKERHIDALE DKIDEEVRRF RETRQKNTNE EDDEVREAMT RARALRSQSE ESASAFSADD LMSIDLAEQM ANDSDD SIS AATNKGRGRG RGRRGGRGQN SASRGGSQRG RADTGLETST RSRNSKTAVS ASRNMSIIDA FKSTRQQPSR NVTTKNY SE VIEVDESDVE EDIFPTTSKT DQRWSSTSSS KIMSQSQVSK GVDFESSEDD DDDPFMNTSS LRRNRRSGGS LEVLFQGP D YKDDDDKGTD YKDDDDK UniProtKB: Double-strand break repair protein MRE11 |
-分子 #2: Nibrin
| 分子 | 名称: Nibrin / タイプ: protein_or_peptide / ID: 2 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 85.073023 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: MWKLLPAAGP AGGEPYRLLT GVEYVVGRKN CAILIENDQS ISRNHAVLTA NFSVTNLSQT DEIPVLTLKD NSKYGTFVNE EKMQNGFSR TLKSGDGITF GVFGSKFRIE YEPLVACSSC LDVSGKTALN QAILQLGGFT VNNWTEECTH LVMVSVKVTI K TICALICG ...文字列: MWKLLPAAGP AGGEPYRLLT GVEYVVGRKN CAILIENDQS ISRNHAVLTA NFSVTNLSQT DEIPVLTLKD NSKYGTFVNE EKMQNGFSR TLKSGDGITF GVFGSKFRIE YEPLVACSSC LDVSGKTALN QAILQLGGFT VNNWTEECTH LVMVSVKVTI K TICALICG RPIVKPEYFT EFLKAVESKK QPPQIESFYP PLDEPSIGSK NVDLSGRQER KQIFKGKTFI FLNAKQHKKL SS AVVFGGG EARLITEENE EEHNFFLAPG TCVVDTGITN SQTLIPDCQK KWIQSIMDML QRQGLRPIPE AEIGLAVIFM TTK NYCDPQ GHPSTGLKTT TPGPSLSQGV SVDEKLMPSA PVNTTTYVAD TESEQADTWD LSERPKEIKV SKMEQKFRML SQDA PTVKE SCKTSSNNNS MVSNTLAKMR IPNYQLSPTK LPSINKSKDR ASQQQQTNSI RNYFQPSTKK RERDEENQEM SSCKS ARIE TSCSLLEQTQ PATPSLWKNK EQHLSENEPV DTNSDNNLFT DTDLKSIVKN SASKSHAAEK LRSNKKREMD DVAIED EVL EQLFKDTKPE LEIDVKVQKQ EEDVNVRKRP RMDIETNDTF SDEAVPESSK ISQENEIGKK RELKEDSLWS AKEISNN DK LQDDSEMLPK KLLLTEFRSL VIKNSTSRNP SGINDDYGQL KNFKKFKKVT YPGAGKLPHI IGGSDLIAHH ARKNTELE E WLRQEMEVQN QHAKEESLAD DLFRYNPYLK RRR UniProtKB: Nibrin |
-分子 #3: MANGANESE (II) ION
| 分子 | 名称: MANGANESE (II) ION / タイプ: ligand / ID: 3 / コピー数: 4 / 式: MN |
|---|---|
| 分子量 | 理論値: 54.938 Da |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.29 mg/mL | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7 構成要素:
詳細: 20 mM HEPES (pH 7.0), 140 mM NaCl, 5 mM MgCl2, 1 mM MnCl2, 0.020 mM ZnCl2, 0.2 mM TCEP, 2 mM ATPgS, plus 0.05 percent beta-OG | |||||||||||||||||||||||||||
| グリッド | モデル: Quantifoil R2/1 / 材質: COPPER / メッシュ: 200 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 7 sec. / 前処理 - 雰囲気: AIR | |||||||||||||||||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 95 % / チャンバー内温度: 288 K / 装置: LEICA EM GP |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 特殊光学系 | エネルギーフィルター - 名称: GIF Bioquantum / エネルギーフィルター - スリット幅: 20 eV |
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / 撮影したグリッド数: 3 / 実像数: 11325 / 平均露光時間: 10.0 sec. / 平均電子線量: 43.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 50.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm 最大 デフォーカス(公称値): 2.8000000000000003 µm 最小 デフォーカス(公称値): 1.0 µm / 倍率(公称値): 13000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー


















 Z
Z Y
Y X
X