[English] 日本語
 Yorodumi
Yorodumi- EMDB-14780: Cryo-EM structure of C-mannosyltransferase CeDPY19, in apo state,... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of C-mannosyltransferase CeDPY19, in apo state, bound to CMT2-Fab and anti-Fab nanobody | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | C-mannosyltransferase / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan / mannosyltransferase activity / nuclear inner membrane / Transferases; Glycosyltransferases; Hexosyltransferases / nervous system development / carbohydrate metabolic process / cell differentiation / endoplasmic reticulum membrane / perinuclear region of cytoplasm / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
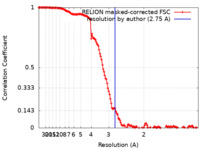
| Method | single particle reconstruction / cryo EM / Resolution: 2.75 Å | |||||||||
 Authors Authors | Bloch JS / Mukherjee S / Irobalieva R / Kossiakoff AA / Goddard-Borger ED / Locher KP | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase. Authors: Joël S Bloch / Alan John / Runyu Mao / Somnath Mukherjee / Jérémy Boilevin / Rossitza N Irobalieva / Tamis Darbre / Nichollas E Scott / Jean-Louis Reymond / Anthony A Kossiakoff / Ethan D ...Authors: Joël S Bloch / Alan John / Runyu Mao / Somnath Mukherjee / Jérémy Boilevin / Rossitza N Irobalieva / Tamis Darbre / Nichollas E Scott / Jean-Louis Reymond / Anthony A Kossiakoff / Ethan D Goddard-Borger / Kaspar P Locher /    Abstract: C-linked glycosylation is essential for the trafficking, folding and function of secretory and transmembrane proteins involved in cellular communication processes. The tryptophan C- ...C-linked glycosylation is essential for the trafficking, folding and function of secretory and transmembrane proteins involved in cellular communication processes. The tryptophan C-mannosyltransferase (CMT) enzymes that install the modification attach a mannose to the first tryptophan of WxxW/C sequons in nascent polypeptide chains by an unknown mechanism. Here, we report cryogenic-electron microscopy structures of Caenorhabditis elegans CMT in four key states: apo, acceptor peptide-bound, donor-substrate analog-bound and as a trapped ternary complex with both peptide and a donor-substrate mimic bound. The structures indicate how the C-mannosylation sequon is recognized by this CMT and its paralogs, and how sequon binding triggers conformational activation of the donor substrate: a process relevant to all glycosyltransferase C superfamily enzymes. Our structural data further indicate that the CMTs adopt an unprecedented electrophilic aromatic substitution mechanism to enable the C-glycosylation of proteins. These results afford opportunities for understanding human disease and therapeutic targeting of specific CMT paralogs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14780.map.gz emd_14780.map.gz | 228.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14780-v30.xml emd-14780-v30.xml emd-14780.xml emd-14780.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14780_fsc.xml emd_14780_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14780.png emd_14780.png | 98.2 KB | ||
| Filedesc metadata |  emd-14780.cif.gz emd-14780.cif.gz | 6.2 KB | ||
| Others |  emd_14780_half_map_1.map.gz emd_14780_half_map_1.map.gz emd_14780_half_map_2.map.gz emd_14780_half_map_2.map.gz | 194.3 MB 194.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14780 http://ftp.pdbj.org/pub/emdb/structures/EMD-14780 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14780 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14780 | HTTPS FTP |
-Validation report
| Summary document |  emd_14780_validation.pdf.gz emd_14780_validation.pdf.gz | 936.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14780_full_validation.pdf.gz emd_14780_full_validation.pdf.gz | 935.7 KB | Display | |
| Data in XML |  emd_14780_validation.xml.gz emd_14780_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_14780_validation.cif.gz emd_14780_validation.cif.gz | 28.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14780 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14780 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14780 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14780 | HTTPS FTP |
-Related structure data
| Related structure data |  7zlhMC  7zlgC  7zliC  7zljC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14780.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14780.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.66 Å | ||||||||||||||||||||||||||||||||||||
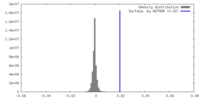
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14780_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
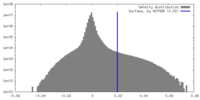
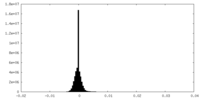
| Density Histograms |
-Half map: #1
| File | emd_14780_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
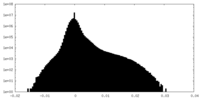
| Density Histograms |
- Sample components
Sample components
-Entire : C-mannosyltransferase CeDPY19, in apo state, bound to CMT2-Fab an...
| Entire | Name: C-mannosyltransferase CeDPY19, in apo state, bound to CMT2-Fab and anti-Fab nanobody |
|---|---|
| Components |
|
-Supramolecule #1: C-mannosyltransferase CeDPY19, in apo state, bound to CMT2-Fab an...
| Supramolecule | Name: C-mannosyltransferase CeDPY19, in apo state, bound to CMT2-Fab and anti-Fab nanobody type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: CMT2-Fab heavy chain
| Macromolecule | Name: CMT2-Fab heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 25.000723 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NISSSSIHWV RQAPGKGLEW VASISSSYGY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARSSSVYWSW WGYSAFDYWG QGTLVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD Y FPEPVTVS ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NISSSSIHWV RQAPGKGLEW VASISSSYGY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARSSSVYWSW WGYSAFDYWG QGTLVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD Y FPEPVTVS WNSGALTSGV HTFPAVLQSS GLYSLSSVVT VPSSSLGTQT YICNVNHKPS NTKVDKKVEP KSCDKTHT |
-Macromolecule #2: Anti-Fab nanobody
| Macromolecule | Name: Anti-Fab nanobody / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 13.390644 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSQVQLQESG GGLVQPGGSL RLSCAASGRT ISRYAMSWFR QAPGKEREFV AVARRSGDGA FYADSVQGRF TVSRDDAKNT VYLQMNSLK PEDTAVYYCA IDSDTFYSGS YDYWGQGTQV TVSS |
-Macromolecule #3: CMT2-Fab light chain
| Macromolecule | Name: CMT2-Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.23875 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQGASEPITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQGASEPITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: C-mannosyltransferase dpy-19
| Macromolecule | Name: C-mannosyltransferase dpy-19 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Glycosyltransferases; Hexosyltransferases |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 80.892602 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKKPKNSPE KSKYSSDTSS SLYSQTWLAS VVIIGLLVGY INYQHVYTLF ENDKHFSHLA DFEREMAYRT EMGLYYSYYK TIINAPSFL EGVQEITHDT VTEHGHEINT LNRFNLYPEV ILAFLYRPFR AFAKSANWQI ELCWQVNRGE LRPVESCEGI G NPHYFYIT ...String: MAKKPKNSPE KSKYSSDTSS SLYSQTWLAS VVIIGLLVGY INYQHVYTLF ENDKHFSHLA DFEREMAYRT EMGLYYSYYK TIINAPSFL EGVQEITHDT VTEHGHEINT LNRFNLYPEV ILAFLYRPFR AFAKSANWQI ELCWQVNRGE LRPVESCEGI G NPHYFYIT GVFIVAGTVA SSIFYLGVLV SDSIFGGFLS VLCFAFNHGE ATRVQWTPPL RESFAFPFII GHIAILTFVI KY KKSGHSM ILLLTSMAVP ALLFWQFTQF AFFTQICSIF LAFSLDLIPF STAKTVIHSH IISFLIGFLL LFGNEMMITA LYF PSILAL GMIIYISPLL SNLKFRPAYV LFLAIIFASI TLGLKIGLSK GLGIEDDAHI FDILRSKFTS FANFHTRLYT CSAE FDFIQ YSTIEKLCGT LLIPLALISL VTFVFNFVKN TNLLWRNSEE IGENGEILYN VVQLCCSTVM AFLIMRLKLF MTPHL CIVA ALFANSKLLG GDRISKTIRV SALVGVIAIL FYRGIPNIRQ QLNVKGEYSN PDQEMLFDWI QHNTKQDAVF AGTMPV MAN VKLTTLRPIV NHPHYEHVGI RERTLKVYSM FSKKPIAEVH KIMKEMGVNY FVFQLMNCSN DERRPECVYR GMWDEED PK NSGRTALCDL WILAANSKDN SRIAPFKIVY NANRNYIVLK ILEDYKDHDG DYKDHDIDYK DDDDK UniProtKB: C-mannosyltransferase dpy-19 |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.8 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)