+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | D. melanogaster 13-protofilament microtubule | |||||||||
 Map data Map data | Cryo-EM map of D. melanogaster microtubule | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cytyoskeleton / microtubules / cytomotive filaments / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationHSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / COPI-mediated anterograde transport / COPI-independent Golgi-to-ER retrograde traffic / COPI-dependent Golgi-to-ER retrograde traffic / Kinesins / astral microtubule / Neutrophil degranulation / lysosome localization / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / structural constituent of cytoskeleton ...HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / COPI-mediated anterograde transport / COPI-independent Golgi-to-ER retrograde traffic / COPI-dependent Golgi-to-ER retrograde traffic / Kinesins / astral microtubule / Neutrophil degranulation / lysosome localization / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / structural constituent of cytoskeleton / microtubule cytoskeleton organization / spindle / mitotic cell cycle / microtubule / hydrolase activity / GTPase activity / centrosome / GTP binding / perinuclear region of cytoplasm / nucleus / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
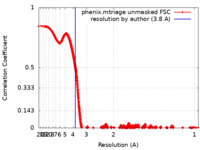
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Wagstaff J / Planelles-Herrero VJ / Derivery E / Lowe J | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics. Authors: James Mark Wagstaff / Vicente José Planelles-Herrero / Grigory Sharov / Aisha Alnami / Frank Kozielski / Emmanuel Derivery / Jan Löwe /  Abstract: Protein filaments are used in myriads of ways to organize other molecules within cells. Some filament-forming proteins couple the hydrolysis of nucleotides to their polymerization cycle, thus ...Protein filaments are used in myriads of ways to organize other molecules within cells. Some filament-forming proteins couple the hydrolysis of nucleotides to their polymerization cycle, thus powering the movement of other molecules. These filaments are termed cytomotive. Only members of the actin and tubulin protein superfamilies are known to form cytomotive filaments. We examined the basis of cytomotivity via structural studies of the polymerization cycles of actin and tubulin homologs from across the tree of life. We analyzed published data and performed structural experiments designed to disentangle functional components of these complex filament systems. Our analysis demonstrates the existence of shared subunit polymerization switches among both cytomotive actins and tubulins, i.e., the conformation of subunits switches upon assembly into filaments. These cytomotive switches can explain filament robustness, by enabling the coupling of kinetic and structural polarities required for cytomotive behaviors and by ensuring that single cytomotive filaments do not fall apart. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14150.map.gz emd_14150.map.gz | 195 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14150-v30.xml emd-14150-v30.xml emd-14150.xml emd-14150.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14150_fsc.xml emd_14150_fsc.xml | 377.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_14150.png emd_14150.png | 222.4 KB | ||
| Filedesc metadata |  emd-14150.cif.gz emd-14150.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14150 http://ftp.pdbj.org/pub/emdb/structures/EMD-14150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14150 | HTTPS FTP |
-Related structure data
| Related structure data |  7qupMC  7qucC  7qudC  7quqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14150.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14150.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of D. melanogaster microtubule | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.102 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Microtubule
| Entire | Name: Microtubule |
|---|---|
| Components |
|
-Supramolecule #1: Microtubule
| Supramolecule | Name: Microtubule / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.170 MDa |
-Macromolecule #1: Tubulin alpha-1 chain
| Macromolecule | Name: Tubulin alpha-1 chain / type: protein_or_peptide / ID: 1 / Number of copies: 39 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.772324 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHEDQ VDPRLIDGKG GGGRPMRECI SIHVGQAGVQ IGNACWELYC LEHGIQPDGQ MPSDKTVGGG DDSFNTFFSE TGAGKHVPR AVFVDLEPTV VDEVRTGTYR QLFHPEQLIT GKEDAANNYA RGHYTIGKEI VDLVLDRIRK LADQCTGLQG F LIFHSFGG ...String: MHHHHHHEDQ VDPRLIDGKG GGGRPMRECI SIHVGQAGVQ IGNACWELYC LEHGIQPDGQ MPSDKTVGGG DDSFNTFFSE TGAGKHVPR AVFVDLEPTV VDEVRTGTYR QLFHPEQLIT GKEDAANNYA RGHYTIGKEI VDLVLDRIRK LADQCTGLQG F LIFHSFGG GTGSGFTSLL MERLSVDYGK KSKLEFAIYP APQVSTAVVE PYNSILTTHT TLEHSDCAFM VDNEAIYDIC RR NLDIERP TYTNLNRLIG QIVSSITASL RFDGALNVDL TEFQTNLVPY PRIHFPLVTY APVISAEKAY HEQLSVAEIT NAC FEPANQ MVKCDPRHGK YMACCMLYRG DVVPKDVNAA IATIKTKRTI QFVDWCPTGF KVGINYQPPT VVPGGDLAKV QRAV CMLSN TTAIAEAWAR LDHKFDLMYA KRAFVHWYVG EGMEEGEFSE AREDLAALEK DYEEVGMDSG DGEGEGAEEY UniProtKB: Tubulin alpha-1 chain |
-Macromolecule #2: Tubulin beta-1 chain
| Macromolecule | Name: Tubulin beta-1 chain / type: protein_or_peptide / ID: 2 / Number of copies: 26 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.681707 KDa |
| Sequence | String: REIVHIQAGQ CGNQIGAKFW EIISDEHGID ATGAYHGDSD LQLERINVYY NEASGGKYVP RAVLVDLEPG TMDSVRSGPF GQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKEAESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP D RIMNTYSV ...String: REIVHIQAGQ CGNQIGAKFW EIISDEHGID ATGAYHGDSD LQLERINVYY NEASGGKYVP RAVLVDLEPG TMDSVRSGPF GQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKEAESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP D RIMNTYSV VPSPKVSDTV VEPYNATLSV HQLVENTDET YCIDNEALYD ICFRTLKLTT PTYGDLNHLV SLTMSGVTTC LR FPGQLNA DLRKLAVNMV PFPRLHFFMP GFAPLTSRGS QQYRALTVPE LTQQMFDAKN MMAACDPRHG RYLTVAAIFR GRM SMKEVD EQMLNIQNKN SSYFVEWIPN NVKTAVCDIP PRGLKMSATF IGNSTAIQEL FKRISEQFTA MFRRKAFLHW YTGE GMDEM EFTEAESNMN DLVSEYQQYQ UniProtKB: Tubulin beta-1 chain |
-Macromolecule #3: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 39 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 39 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 26 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.9 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller