[English] 日本語
 Yorodumi
Yorodumi- EMDB-13745: RNA polymerase elongation complex in more-swiveled conformation -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA polymerase elongation complex in more-swiveled conformation | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | transcription elongation / cryo-EM / TRANSCRIPTION | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / regulation of DNA-templated transcription initiation / bacterial-type flagellum assembly / bacterial-type flagellum-dependent cell motility / nitrate assimilation / DNA-directed RNA polymerase complex ...RNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / regulation of DNA-templated transcription initiation / bacterial-type flagellum assembly / bacterial-type flagellum-dependent cell motility / nitrate assimilation / DNA-directed RNA polymerase complex / transcription elongation factor complex / regulation of DNA-templated transcription elongation / DNA-templated transcription initiation / transcription antitermination / cell motility / ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity / DNA-directed RNA polymerase / response to heat / protein-containing complex assembly / intracellular iron ion homeostasis / protein dimerization activity / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | ||||||||||||
 Authors Authors | Zhu C / Guo X | ||||||||||||
| Funding support |  France, 3 items France, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Transcription factors modulate RNA polymerase conformational equilibrium. Authors: Chengjin Zhu / Xieyang Guo / Philippe Dumas / Maria Takacs / Mo'men Abdelkareem / Arnaud Vanden Broeck / Charlotte Saint-André / Gabor Papai / Corinne Crucifix / Julio Ortiz / Albert Weixlbaumer /    Abstract: RNA polymerase (RNAP) frequently pauses during the transcription of DNA to RNA to regulate gene expression. Transcription factors NusA and NusG modulate pausing, have opposing roles, but can bind ...RNA polymerase (RNAP) frequently pauses during the transcription of DNA to RNA to regulate gene expression. Transcription factors NusA and NusG modulate pausing, have opposing roles, but can bind RNAP simultaneously. Here we report cryo-EM reconstructions of Escherichia coli RNAP bound to NusG, or NusA, or both. RNAP conformational changes, referred to as swivelling, correlate with transcriptional pausing. NusA facilitates RNAP swivelling to further increase pausing, while NusG counteracts this role. Their structural effects are consistent with biochemical results on two categories of transcriptional pauses. In addition, the structures suggest a cooperative mechanism of NusA and NusG during Rho-mediated transcription termination. Our results provide a structural rationale for the stochastic nature of pausing and termination and how NusA and NusG can modulate it. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13745.map.gz emd_13745.map.gz | 79.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13745-v30.xml emd-13745-v30.xml emd-13745.xml emd-13745.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13745.png emd_13745.png | 146.9 KB | ||
| Filedesc metadata |  emd-13745.cif.gz emd-13745.cif.gz | 7.9 KB | ||
| Others |  emd_13745_additional_1.map.gz emd_13745_additional_1.map.gz emd_13745_additional_2.map.gz emd_13745_additional_2.map.gz | 41.9 MB 75.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13745 http://ftp.pdbj.org/pub/emdb/structures/EMD-13745 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13745 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13745 | HTTPS FTP |
-Validation report
| Summary document |  emd_13745_validation.pdf.gz emd_13745_validation.pdf.gz | 500.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13745_full_validation.pdf.gz emd_13745_full_validation.pdf.gz | 500 KB | Display | |
| Data in XML |  emd_13745_validation.xml.gz emd_13745_validation.xml.gz | 6.4 KB | Display | |
| Data in CIF |  emd_13745_validation.cif.gz emd_13745_validation.cif.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13745 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13745 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13745 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13745 | HTTPS FTP |
-Related structure data
| Related structure data |  7q0jMC  7py0C  7py1C  7py3C  7py5C  7py6C  7py7C  7py8C  7pyjC  7pykC  7q0kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13745.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13745.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_13745_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
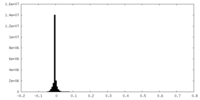
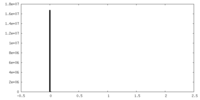
| Density Histograms |
-Additional map: #2
| File | emd_13745_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
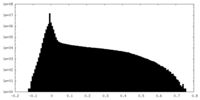
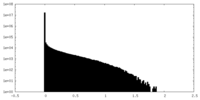
| Density Histograms |
- Sample components
Sample components
+Entire : RNA polymerase elongation complex in more-swiveled conformation
+Supramolecule #1: RNA polymerase elongation complex in more-swiveled conformation
+Macromolecule #1: DNA-directed RNA polymerase subunit alpha
+Macromolecule #2: DNA-directed RNA polymerase subunit beta
+Macromolecule #3: DNA-directed RNA polymerase subunit beta'
+Macromolecule #4: DNA-directed RNA polymerase subunit omega
+Macromolecule #5: ntDNA
+Macromolecule #7: tDNA
+Macromolecule #6: RNA
+Macromolecule #8: ZINC ION
+Macromolecule #9: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 67666 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)