+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13733 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | HBc-P5T in complex with X-100 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HBc-P5T (low secretion phenotype) in complex with Triton X-100 / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule-dependent intracellular transport of viral material towards nucleus / T=4 icosahedral viral capsid / viral penetration into host nucleus / host cell / host cell cytoplasm / symbiont entry into host cell / structural molecule activity / DNA binding / RNA binding Similarity search - Function | |||||||||
| Biological species |  Hepatitis B virus ayw/France/Tiollais/1979 / Hepatitis B virus ayw/France/Tiollais/1979 /  Hepatitis B virus genotype D subtype ayw (isolate France/Tiollais/1979) Hepatitis B virus genotype D subtype ayw (isolate France/Tiollais/1979) | |||||||||
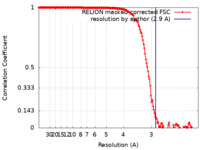
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Makbul C / Boettcher B | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Viruses / Year: 2021 Journal: Viruses / Year: 2021Title: Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97. Authors: Cihan Makbul / Christian Kraft / Matthias Grießmann / Tim Rasmussen / Kilian Katzenberger / Melina Lappe / Paul Pfarr / Cato Stoffer / Mara Stöhr / Anna-Maria Wandinger / Bettina Böttcher /  Abstract: (1) Background: During maturation of the Hepatitis B virus, a viral polymerase inside the capsid transcribes a pre-genomic RNA into a partly double stranded DNA-genome. This is followed by ...(1) Background: During maturation of the Hepatitis B virus, a viral polymerase inside the capsid transcribes a pre-genomic RNA into a partly double stranded DNA-genome. This is followed by envelopment with surface proteins inserted into a membrane. Envelopment is hypothetically regulated by a structural signal that reports the maturation state of the genome. NMR data suggest that such a signal can be mimicked by the binding of the detergent Triton X 100 to hydrophobic pockets in the capsid spikes. (2) Methods: We have used electron cryo-microscopy and image processing to elucidate the structural changes that are concomitant with the binding of Triton X 100. (3) Results: Our maps show that Triton X 100 binds with its hydrophobic head group inside the pocket. The hydrophilic tail delineates the outside of the spike and is coordinated via Lys-96. The binding of Triton X 100 changes the rotamer conformation of Phe-97 in helix 4, which enables a π-stacking interaction with Trp-62 in helix 3. Similar changes occur in mutants with low secretion phenotypes (P5T and L60V) and in a mutant with a pre-mature secretion phenotype (F97L). (4) Conclusion: Binding of Triton X 100 is unlikely to mimic structural maturation because mutants with different secretion phenotypes show similar structural responses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13733.map.gz emd_13733.map.gz | 228.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13733-v30.xml emd-13733-v30.xml emd-13733.xml emd-13733.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13733_fsc.xml emd_13733_fsc.xml | 15.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_13733.png emd_13733.png | 221.3 KB | ||
| Filedesc metadata |  emd-13733.cif.gz emd-13733.cif.gz | 5.4 KB | ||
| Others |  emd_13733_half_map_1.map.gz emd_13733_half_map_1.map.gz emd_13733_half_map_2.map.gz emd_13733_half_map_2.map.gz | 260 MB 260 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13733 http://ftp.pdbj.org/pub/emdb/structures/EMD-13733 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13733 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13733 | HTTPS FTP |
-Validation report
| Summary document |  emd_13733_validation.pdf.gz emd_13733_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13733_full_validation.pdf.gz emd_13733_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_13733_validation.xml.gz emd_13733_validation.xml.gz | 23.6 KB | Display | |
| Data in CIF |  emd_13733_validation.cif.gz emd_13733_validation.cif.gz | 30.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13733 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13733 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13733 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13733 | HTTPS FTP |
-Related structure data
| Related structure data |  7pzmMC  7pz9C  7pziC  7pzkC  7pzlC  7pznC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13733.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13733.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0635 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_13733_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13733_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HBc-P5T-TX100
| Entire | Name: HBc-P5T-TX100 |
|---|---|
| Components |
|
-Supramolecule #1: HBc-P5T-TX100
| Supramolecule | Name: HBc-P5T-TX100 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Hepatitis B virus ayw/France/Tiollais/1979 Hepatitis B virus ayw/France/Tiollais/1979 |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Hepatitis B virus genotype D subtype ayw (isolate France/Tiollais/1979) Hepatitis B virus genotype D subtype ayw (isolate France/Tiollais/1979)Strain: isolate France/Tiollais/1979 |
| Molecular weight | Theoretical: 21.150205 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDIDTYKEFG ATVELLSFLP SDFFPSVRDL LDTASALYRE ALESPEHCSP HHTALRQAIL CWGELMTLAT WVGVNLEDPA SRDLVVSYV NTNMGLKFRQ LLWFHISCLT FGRETVIEYL VSFGVWIRTP PAYRPPNAPI LSTLPETTVV RRRGRSPRRR T PSPRRRRS QSPRRRRSQS RESQC UniProtKB: Capsid protein |
-Macromolecule #2: FRAGMENT OF TRITON X-100
| Macromolecule | Name: FRAGMENT OF TRITON X-100 / type: ligand / ID: 2 / Number of copies: 4 / Formula: TRT |
|---|---|
| Molecular weight | Theoretical: 352.508 Da |
| Chemical component information |  ChemComp-TRT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.029 kPa |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)