[English] 日本語
 Yorodumi
Yorodumi- EMDB-13268: Streptococcus pneumoniae choline importer LicB in lipid nanodiscs -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13268 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Streptococcus pneumoniae choline importer LicB in lipid nanodiscs | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  EamA domain / EamA-like transporter family / membrane => GO:0016020 / LicB protein EamA domain / EamA-like transporter family / membrane => GO:0016020 / LicB protein Function and homology information Function and homology information | |||||||||
| Biological species |   Streptococcus pneumoniae TIGR4 (bacteria) / synthetic construct (others) Streptococcus pneumoniae TIGR4 (bacteria) / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.75 Å cryo EM / Resolution: 3.75 Å | |||||||||
 Authors Authors | Perez C / Baerland N | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Mechanistic basis of choline import involved in teichoic acids and lipopolysaccharide modification. Authors: Natalie Bärland / Anne-Stéphanie Rueff / Gonzalo Cebrero / Cedric A J Hutter / Markus A Seeger / Jan-Willem Veening / Camilo Perez /  Abstract: Phosphocholine molecules decorating bacterial cell wall teichoic acids and outer-membrane lipopolysaccharide have fundamental roles in adhesion to host cells, immune evasion, and persistence. ...Phosphocholine molecules decorating bacterial cell wall teichoic acids and outer-membrane lipopolysaccharide have fundamental roles in adhesion to host cells, immune evasion, and persistence. Bacteria carrying the operon that performs phosphocholine decoration synthesize phosphocholine after uptake of the choline precursor by LicB, a conserved transporter among divergent species. is a prominent pathogen where phosphocholine decoration plays a fundamental role in virulence. Here, we present cryo-electron microscopy and crystal structures of LicB, revealing distinct conformational states and describing architectural and mechanistic elements essential to choline import. Together with in vitro and in vivo functional characterization, we found that LicB displays proton-coupled import activity and promiscuous selectivity involved in adaptation to choline deprivation conditions, and describe LicB inhibition by synthetic nanobodies (sybodies). Our results provide previously unknown insights into the molecular mechanism of a key transporter involved in bacterial pathogenesis and establish a basis for inhibition of the phosphocholine modification pathway across bacterial phyla. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: Mechanistic basis of choline import involved in teichoic acids and lipopolysaccharide modification Authors: Barland N / Rueff AS / Cebrero G / Hutter CA / Seeger MA / Veening JW / Perez C | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13268.map.gz emd_13268.map.gz | 203.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13268-v30.xml emd-13268-v30.xml emd-13268.xml emd-13268.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13268.png emd_13268.png | 59.9 KB | ||
| Masks |  emd_13268_msk_1.map emd_13268_msk_1.map | 216 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13268 http://ftp.pdbj.org/pub/emdb/structures/EMD-13268 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13268 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13268 | HTTPS FTP |
-Related structure data
| Related structure data |  7pafMC  7b0kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13268.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13268.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.878 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13268_msk_1.map emd_13268_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
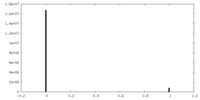
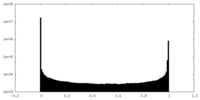
| Density Histograms |
- Sample components
Sample components
-Entire : Choline importer in nanodiscs bound to nanobody
| Entire | Name: Choline importer in nanodiscs bound to nanobody |
|---|---|
| Components |
|
-Supramolecule #1: Choline importer in nanodiscs bound to nanobody
| Supramolecule | Name: Choline importer in nanodiscs bound to nanobody / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Streptococcus pneumoniae TIGR4 (bacteria) Streptococcus pneumoniae TIGR4 (bacteria) |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Molecular weight | Experimental: 260 KDa |
-Macromolecule #1: Nanobody
| Macromolecule | Name: Nanobody / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 16.802365 KDa |
| Recombinant expression | Organism:   Escherichia coli MC1061 (bacteria) Escherichia coli MC1061 (bacteria) |
| Sequence | String: GSSSQVQLVE SGGGSVQAGG SLRLSCAASG TIHAIGYLGW FRQAPGKERE GVAALTTYDG WTYYADSVKG RFTVSLDNAK NTVYLQMNS LKPEDTALYY CAAADDGWMF PLYHNHYEYW GQGTQVTVSA GRAGEQKLIS EEDLNSAVDH HHHHH |
-Macromolecule #2: LicB protein
| Macromolecule | Name: LicB protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Streptococcus pneumoniae TIGR4 (bacteria) / Strain: ATCC BAA-334 / TIGR4 Streptococcus pneumoniae TIGR4 (bacteria) / Strain: ATCC BAA-334 / TIGR4 |
| Molecular weight | Theoretical: 31.647631 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MKSKNGVPFG LLSGIFWGLG LTVSAYIFSI FTDLSPFVVA ATHDFLSIFI LLAFLLVKEG KVRLSIFLNI RNVSVIIGAL LAGPIGMQA NLYAVKYIGS SLASSVSAIY PAISVLLAFF FLKHKISKNT VFGIVLIIGG IIAQTYKVEQ VNSFYIGILC A LVCAIAWG ...String: MKSKNGVPFG LLSGIFWGLG LTVSAYIFSI FTDLSPFVVA ATHDFLSIFI LLAFLLVKEG KVRLSIFLNI RNVSVIIGAL LAGPIGMQA NLYAVKYIGS SLASSVSAIY PAISVLLAFF FLKHKISKNT VFGIVLIIGG IIAQTYKVEQ VNSFYIGILC A LVCAIAWG SESVLSSFAM ESELSEIEAL LIRQVTSFLS YLVIVLFSHQ SFTAVANGQL LGLMIVFAAF DMISYLAYYI AI NRLQPAK ATGLNVSYVV WTVLFAVVFL GAPLDMLTIM TSLVVIAGVY IIIKE |
-Macromolecule #3: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
| Macromolecule | Name: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE type: ligand / ID: 3 / Number of copies: 1 / Formula: PGT |
|---|---|
| Molecular weight | Theoretical: 751.023 Da |
| Chemical component information |  ChemComp-PGT: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.0 µm Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
- Image processing
Image processing
| CTF correction | Software - Name: cryoSPARC (ver. v3.2.0) |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: cryoSPARC (ver. v3.2.0) |
| Final 3D classification | Number classes: 4 / Software - Name: cryoSPARC (ver. v3.2.0) |
| Final angle assignment | Type: OTHER / Software - Name: cryoSPARC (ver. v3.2.0) |
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic ) / Resolution.type: BY AUTHOR / Resolution: 3.75 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. v3.2.0) / Number images used: 78649 ) / Resolution.type: BY AUTHOR / Resolution: 3.75 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. v3.2.0) / Number images used: 78649 |
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|---|
| Output model |  PDB-7paf: |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X