+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13171 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Signal Peptidase Complex Paralog A (SPC-A) | |||||||||
 Map data Map data | Amphipol has been largely masked out | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Endoplasmic Reticulum / Signal peptide / Serine protease / Membrane complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsignal peptidase complex / signal peptidase I / signal peptide processing / protein targeting to ER / Synthesis, secretion, and inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) / virion assembly / Maturation of hRSV A proteins / SRP-dependent cotranslational protein targeting to membrane / Synthesis, secretion, and deacylation of Ghrelin / response to virus ...signal peptidase complex / signal peptidase I / signal peptide processing / protein targeting to ER / Synthesis, secretion, and inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) / virion assembly / Maturation of hRSV A proteins / SRP-dependent cotranslational protein targeting to membrane / Synthesis, secretion, and deacylation of Ghrelin / response to virus / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / peptidase activity / viral protein processing / serine-type endopeptidase activity / endoplasmic reticulum membrane / endoplasmic reticulum / proteolysis Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
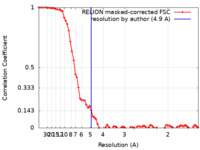
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Liaci AM / Foerster F | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage. Authors: A Manuel Liaci / Barbara Steigenberger / Paulo Cesar Telles de Souza / Sem Tamara / Mariska Gröllers-Mulderij / Patrick Ogrissek / Siewert J Marrink / Richard A Scheltema / Friedrich Förster /    Abstract: The signal peptidase complex (SPC) is an essential membrane complex in the endoplasmic reticulum (ER), where it removes signal peptides (SPs) from a large variety of secretory pre-proteins with ...The signal peptidase complex (SPC) is an essential membrane complex in the endoplasmic reticulum (ER), where it removes signal peptides (SPs) from a large variety of secretory pre-proteins with exquisite specificity. Although the determinants of this process have been established empirically, the molecular details of SP recognition and removal remain elusive. Here, we show that the human SPC exists in two functional paralogs with distinct proteolytic subunits. We determined the atomic structures of both paralogs using electron cryo-microscopy and structural proteomics. The active site is formed by a catalytic triad and abuts the ER membrane, where a transmembrane window collectively formed by all subunits locally thins the bilayer. Molecular dynamics simulations indicate that this unique architecture generates specificity for SPs based on the length of their hydrophobic segments. #1:  Journal: BioRxiv / Year: 2020 Journal: BioRxiv / Year: 2020Title: Structure of the Human Signal Peptidase Complex Reveals the Determinants for Signal Peptide Cleavage Authors: Liaci AM / Steigenberger B / Tamara S / Telles de Souza PC / Grollers-Mulderij M / Ogrissek P / Marrik SJ / Scheltema RA / Foerster F | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13171.map.gz emd_13171.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13171-v30.xml emd-13171-v30.xml emd-13171.xml emd-13171.xml | 31 KB 31 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13171_fsc.xml emd_13171_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_13171.png emd_13171.png | 104.7 KB | ||
| Masks |  emd_13171_msk_1.map emd_13171_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13171.cif.gz emd-13171.cif.gz | 7.5 KB | ||
| Others |  emd_13171_half_map_1.map.gz emd_13171_half_map_1.map.gz emd_13171_half_map_2.map.gz emd_13171_half_map_2.map.gz | 28.3 MB 28.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13171 http://ftp.pdbj.org/pub/emdb/structures/EMD-13171 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13171 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13171 | HTTPS FTP |
-Validation report
| Summary document |  emd_13171_validation.pdf.gz emd_13171_validation.pdf.gz | 966.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13171_full_validation.pdf.gz emd_13171_full_validation.pdf.gz | 966.1 KB | Display | |
| Data in XML |  emd_13171_validation.xml.gz emd_13171_validation.xml.gz | 12.7 KB | Display | |
| Data in CIF |  emd_13171_validation.cif.gz emd_13171_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13171 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13171 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13171 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13171 | HTTPS FTP |
-Related structure data
| Related structure data |  7p2pMC  7p2qC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13171.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13171.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Amphipol has been largely masked out | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13171_msk_1.map emd_13171_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13171_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13171_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Signal peptidase complex paralog A (SPC-A)
| Entire | Name: Signal peptidase complex paralog A (SPC-A) |
|---|---|
| Components |
|
-Supramolecule #1: Signal peptidase complex paralog A (SPC-A)
| Supramolecule | Name: Signal peptidase complex paralog A (SPC-A) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organelle: Endoplasmic Reticulum / Location in cell: Endoplasmic Reticulum Homo sapiens (human) / Organelle: Endoplasmic Reticulum / Location in cell: Endoplasmic Reticulum |
| Molecular weight | Theoretical: 20.3 KDa |
-Supramolecule #2: Signal peptidase complex catalytic subunit A (SEC11A)
| Supramolecule | Name: Signal peptidase complex catalytic subunit A (SEC11A) / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organelle: Endoplasmic Reticulum / Location in cell: Endoplasmic Reticulum Homo sapiens (human) / Organelle: Endoplasmic Reticulum / Location in cell: Endoplasmic Reticulum |
-Supramolecule #3: Signal peptidase complex subunit 3 (SPC22/23)
| Supramolecule | Name: Signal peptidase complex subunit 3 (SPC22/23) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organelle: Endoplasmic Reticulum / Location in cell: Endoplasmic Reticulum Homo sapiens (human) / Organelle: Endoplasmic Reticulum / Location in cell: Endoplasmic Reticulum |
-Supramolecule #4: Signal peptidase complex subunit 2 (SPC25)
| Supramolecule | Name: Signal peptidase complex subunit 2 (SPC25) / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organelle: Endoplasmic Reticulum / Location in cell: Endoplasmic Reticulum Homo sapiens (human) / Organelle: Endoplasmic Reticulum / Location in cell: Endoplasmic Reticulum |
-Supramolecule #5: Signal peptidase complex subunit 1 (SPC12)
| Supramolecule | Name: Signal peptidase complex subunit 1 (SPC12) / type: complex / ID: 5 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organelle: Endoplasmic Reticulum / Location in cell: Endoplasmic Reticulum Homo sapiens (human) / Organelle: Endoplasmic Reticulum / Location in cell: Endoplasmic Reticulum |
-Macromolecule #1: Signal peptidase complex catalytic subunit SEC11A
| Macromolecule | Name: Signal peptidase complex catalytic subunit SEC11A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: signal peptidase I |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.376088 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PMLSLDFLDD VRRMNKRQLY YQVLNFGMIV SSALMIWKGL MVITGSESPI VVVLSGSMEP AFHRGDLLFL TNRVEDPIRV GEIVVFRIE GREIPIVHRV LKIHEKQNGH IKFLTKGDNN AVDDRGLYKQ GQHWLEKKDV VGRARGFVPY IGIVTILMND Y PKFKYAVL ...String: PMLSLDFLDD VRRMNKRQLY YQVLNFGMIV SSALMIWKGL MVITGSESPI VVVLSGSMEP AFHRGDLLFL TNRVEDPIRV GEIVVFRIE GREIPIVHRV LKIHEKQNGH IKFLTKGDNN AVDDRGLYKQ GQHWLEKKDV VGRARGFVPY IGIVTILMND Y PKFKYAVL FLLGLFVLVH REGGSPGGSG GGSAENLYFQ UniProtKB: Signal peptidase complex catalytic subunit SEC11A |
-Macromolecule #2: Signal peptidase complex subunit 3
| Macromolecule | Name: Signal peptidase complex subunit 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on peptide bonds (peptidases) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.348457 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PMNTVLSRAN SLFAFSLSVM AALTFGCFIT TAFKDRSVPV RLHVSRIMLK NVEDFTGPRE RSDLGFITFD ITADLENIFD WNVKQLFLY LSAEYSTKNN ALNQVVLWDK IVLRGDNPKL LLKDMKTKYF FFDDGNGLKG NRNVTLTLSW NVVPNAGILP L VTGSGHVS ...String: PMNTVLSRAN SLFAFSLSVM AALTFGCFIT TAFKDRSVPV RLHVSRIMLK NVEDFTGPRE RSDLGFITFD ITADLENIFD WNVKQLFLY LSAEYSTKNN ALNQVVLWDK IVLRGDNPKL LLKDMKTKYF FFDDGNGLKG NRNVTLTLSW NVVPNAGILP L VTGSGHVS VPFPDTYEIT KSYGSGEGRG SLLTCGDVEE NPG UniProtKB: Signal peptidase complex subunit 3 |
-Macromolecule #3: Signal peptidase complex subunit 2
| Macromolecule | Name: Signal peptidase complex subunit 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on peptide bonds (peptidases) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.215236 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAAAVQGGR SGGSGGCSGA GGASNCGTGS GRSGLLDKWK IDDKPVKIDK WDGSAVKNSL DDSAKKVLLE KYKYVENFGL IDGRLTICT ISCFFAIVAL IWDYMHPFPE SKPVLALCVI SYFVMMGILT IYTSYKEKSI FLVAHRKDPT GMDPDDIWQL S SSLKRFDD ...String: MAAAAVQGGR SGGSGGCSGA GGASNCGTGS GRSGLLDKWK IDDKPVKIDK WDGSAVKNSL DDSAKKVLLE KYKYVENFGL IDGRLTICT ISCFFAIVAL IWDYMHPFPE SKPVLALCVI SYFVMMGILT IYTSYKEKSI FLVAHRKDPT GMDPDDIWQL S SSLKRFDD KYTLKLTFIS GRTKQQREAE FTKSIAKFFD HSGTLVMDAY EPEISRLHDS LAIERKIKGS GQCTNYALLK LA GDVESNP G UniProtKB: Signal peptidase complex subunit 2 |
-Macromolecule #4: Signal peptidase complex subunit 1
| Macromolecule | Name: Signal peptidase complex subunit 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on peptide bonds (peptidases) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.488445 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PMARGGDTGC TGPSETSASG AAAIALPGLE GPATDAQCQT LPLTVLKSRS PSPRSLPPAL SCPPPQPAML EHLSSLPTQM DYKGQKLAE QMFQGIILFS AIVGFIYGYV AEQFGWTVYI VMAGFAFSCL LTLPPWPIYR RHPLKWLPVQ ESSTDDKKPG E RKIKRHAK NNGSGATNFS LLKQAGDVEE NPG UniProtKB: Signal peptidase complex subunit 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation #1
Sample preparation #1
| Preparation ID | 1 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Concentration | 0.5 mg/mL | |||||||||||||||
| Buffer | pH: 7.8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Film type ID: 1 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 4s with blot force 0 before plunging.. | |||||||||||||||
| Details | grid condition 1 (frozen without fluorinated fos-choline-8) |
- Sample preparation #2
Sample preparation #2
| Preparation ID | 2 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Concentration | 4 mg/mL | |||||||||||||||
| Buffer | pH: 7.8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Film type ID: 1 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 4s with blot force 0 before plunging.. | |||||||||||||||
| Details | grid condition 2 (frozen with 1.5 mM fluorinated fos-choline-8) |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 93.0 K / Max: 93.0 K |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Details | 200 kV Talos Arctica at Utrecht University, the Netherlands. Same settings were used for both grid conditions. |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #0 - Detector mode: COUNTING / #0 - Number grids imaged: 1 / #0 - Number real images: 5203 / #0 - Average exposure time: 9.0 sec. / #0 - Average electron dose: 63.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #1 - Detector mode: COUNTING / #1 - Digitization - Dimensions - Width: 3838 pixel / #1 - Digitization - Dimensions - Height: 3710 pixel / #1 - Number grids imaged: 1 / #1 - Number real images: 2083 / #1 - Average exposure time: 10.2 sec. / #1 - Average electron dose: 63.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 30.0 µm / Calibrated defocus max: 4.8 µm / Calibrated defocus min: 0.3 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7p2p: |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)